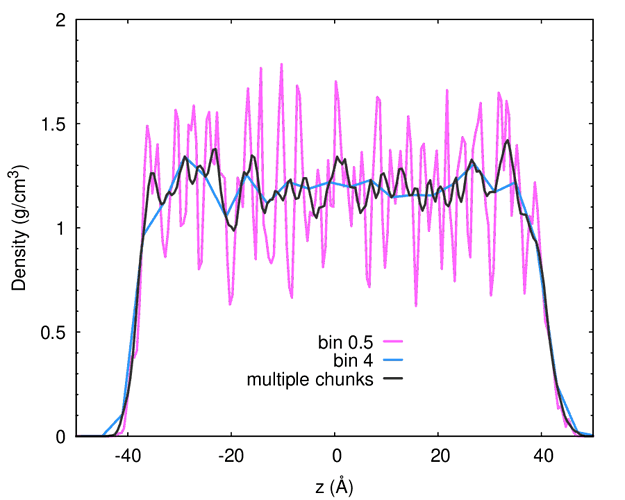
The task is to compute a density profile for a slab, making it less noisy by using a sufficiently large bin. The problem is that the larger the bin, the coarser the profile is. My solution is to compute multiple chunks, whose number is bin/resolution, in this case, I want a spacing of 0.5 A and a bin size of 4 A, hence I need 8 computes.
The problem is that I haven’t found an elegant fix to combine the chunks into a single output, so my ugly solution is to define an identical number of fix ave/chunk and then combine the output files into a single one at the end of the calculation. Here the relevant snippet:
compute c2 all chunk/atom bin/1d z -55.5 4 units box
compute c3 all chunk/atom bin/1d z -56.0 4 units box
compute c4 all chunk/atom bin/1d z -56.5 4 units box
compute c5 all chunk/atom bin/1d z -57.0 4 units box
compute c6 all chunk/atom bin/1d z -57.5 4 units box
compute c7 all chunk/atom bin/1d z -58.0 4 units box
compute c8 all chunk/atom bin/1d z -58.5 4 units box
# average every 1 frame, repeat 500 times, print only once at last timestep.
fix f1 all ave/chunk 1 500 2502 c1 density/mass ave running file ${run}.out1
fix f2 all ave/chunk 1 500 2502 c2 density/mass ave running file ${run}.out2
fix f3 all ave/chunk 1 500 2502 c3 density/mass ave running file ${run}.out3
fix f4 all ave/chunk 1 500 2502 c4 density/mass ave running file ${run}.out4
fix f5 all ave/chunk 1 500 2502 c5 density/mass ave running file ${run}.out5
fix f6 all ave/chunk 1 500 2502 c6 density/mass ave running file ${run}.out6
fix f7 all ave/chunk 1 500 2502 c7 density/mass ave running file ${run}.out7
fix f8 all ave/chunk 1 500 2502 c8 density/mass ave running file ${run}.out8
# Output.
thermo 1
thermo_style custom step cpu
thermo_modify flush yes
rerun ../${run}.dcd first 2002 dump x y z box yes format molfile dcd /usr/local/lib/vmd/plugins/LINUXAMD64/molfile
# Merge all output files:
shell bash merge.sh ${run}.out1 ${run}.out2 ${run}.out3 ${run}.out4 ${run}.out5 ${run}.out6 ${run}.out7 ${run}.out8 > ${run}.out
# Where the script merge.sh is as follows:
## Extract the column 2 and 4 from a number of files.
#awk '$1>0{print $2,$4}' $@ | sort -n
## hide my crimes.
#rm -f $@
This is the result. For comparison, you can see the outcome of using a single bin size, in this case with spacing 0.5 (noisy profile) and 4 Angstroms (coarse profile).
Any idea?