Dear all
Is there any way to twist the whole simulation box like this? Periodic boundary conditions are applied on the twisting axis while other boundary conditions could be any type. Thanks very much and any recommendations are welcome.
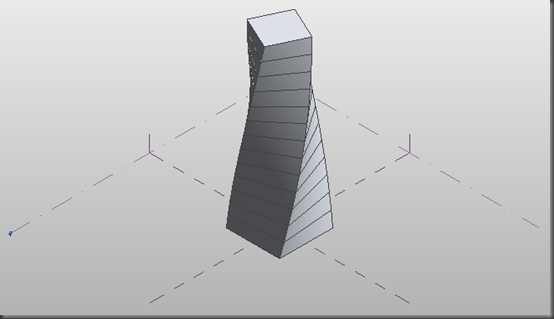
Dear all
Is there any way to twist the whole simulation box like this? Periodic boundary conditions are applied on the twisting axis while other boundary conditions could be any type. Thanks very much and any recommendations are welcome.

Dear all
Is there any way to twist the whole simulation box like this? Periodic
boundary conditions are applied on the twisting axis while other boundary
conditions could be any type. Thanks very much and any recommendations are
welcome.
not as such. do you need to have the degree of twisting changed during the
simulation or is it sufficient to be constant?
in the latter case, your problem simply becomes a problem of creating the
initial coordinates, which should be doable with LAMMPS.
axel.

Dear Axel
Unfortunately, I need it to change constantly through the whole simulation. So, there is no such command to achieve it?
Best regards
Rita

Dear Axel
Unfortunately, I need it to change constantly through the whole
simulation. So, there is no such command to achieve it?
no, there is only the option to tilt a triclinic box.
i am wondering, though, whether it should not be possible to project your
problem into that kind of system.

My original goal is to twist a tube which is made up by DNA origami. Degrees of freedom in my simulation are relatively large, and the simulation time are very long because the twisting velocity is extremely small (it should not be increased because with larger velocity, the DNA origami corrupt before being twisted), so I could not model the tube with the length/radius ratio larger than 10 (the radius could not be reduced because of the stiffness of DNA origami), but my professor insists if the length/radius ratio is smaller than 10, boundary effects will not be eliminated. So I am quite confused about how to meet my professor’s requirements.

that sounds like you should have a chat with your adviser then. after all,
it is the job of an adviser to give advice or at the very least point you
in the right direction so that you can figure it out by yourself. keep in
mind that when doing research things don't always work out as expected; if
people would already know the answer, it would not be research. thus there
is no point in agonizing over something that cannot be done exactly in the
way as intended, but at the same time, you should verify that you have good
arguments to support this claim. perhaps you overlooked something that
makes your simulations run inefficiently. i also am not convinced that the
kind of boundary conditions you ask for, would be applicable in your
system.
axel.
Dear Axel
Thanks very much for your suggestions and encouragements, I will talk to my adviser and find an solution. If there is any progress that will be helpful for others, I will reply to you so that others could see.
Best regards
Rita
Comment: Unfortunately even if this feature were available in
LAMMPS, this kind of boundary condition does not describe a system
which is periodic in all 3 dimensions (x,y,z). These means that it
would be almost impossible to add explicit water to your simulations,
regardless of what software you used (LAMMPS, GROMACS, NAMD)... At
least I can't think of an easy way.
--- more pontification ---- (feel free to skip)
...This is an interesting idea. For one thing, many helical
polymers, the number of monomers per twist is usually never exactly an
integer. (...even when they in the relaxed state and are not
experiencing twist forces. For DNA it is either ~5.2 or ~10.5
base-pairs per twist, depending on symmetry conventions.) But it's
usually close enough that people fudge it (especially if you are
willing to simulate a longer polymer spanning several multiples of the
twist period. (I think I've seen DNA simulations with 21 base-pairs,
for example). People either do that, or they simulate a long polymer
which is not periodic. Yihua, if you have enough computational
resources, -that- might be the best option.
If you want to be rigorous, one could imagine some kind of
equilibrium ensemble using Yihua's boundary conditions, that has
constant Number-of-particles, Tension, Torsion, and Temperature
(...the "NTTT ensemble"?)
However I doubt it is worth undertaking the painful process of
editing the code to implement this kind of boundary in an MD program.
(Feel free to chime-in if you disagree) Yihua I strongly suspect that
there are other ways to answer the question you are really interested
in. Peel free to let us know if you find a good work-around.
Nice picture, by the way.
Andrew