Hi list,
I used MAPS to build structure of a glycerol, then mininized the structure.
After that I took the restart file and run NVT at 300K. All steps are
simple. However, I got lots of “nan” in the output when running NVT and
could not explain why.
It’s good be great if you could help me to solve the problem.
Thanks,
Ly Le, Univ of Utah
in.test (1.3 KB)
data.glycerol (4.89 KB)
log.lammps (9.66 KB)
restart_glycerol.minimized (5.48 KB)
Ly Le,
You did not create glycerol. You created something a-chemical (see attached). You have -C-H-H bonds instead of -C-O-H or something (see attached).
Pieter
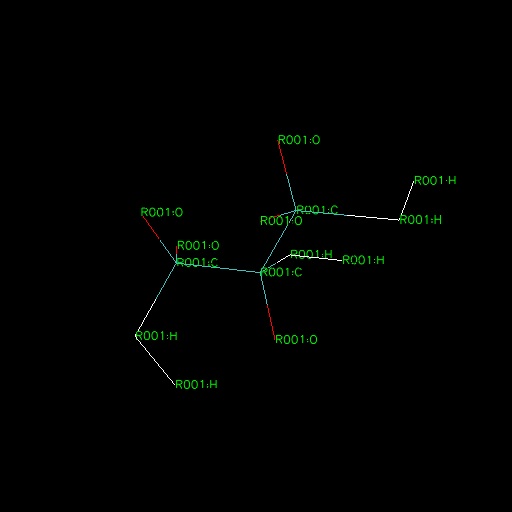
Ly Le,
I retract my earlier remark. My lammps2pdb.pl script does not interpret the mass indices. I just noticed they are out of order in your .data file.
Pieter
There are many ways to get NaNs. Is your starting structure low-energy
and low-pressure. Else you may have large forces. Small enough timesteps?
Etc, etc. Try to figure out when it is going bad by printing thermo every step,
then figure out why.
Steve
Hi list,
I used MAPS to build structure of a glycerol, then mininized the
structure.
After that I took the restart file and run NVT at 300K. All steps are
simple. However, I got lots of "nan" in the output when running NVT and
could not explain why.
how about calling a 10fs timestep more than a bit optimistic?
try with 1fs.
cheers,
axel.
