Dear all,
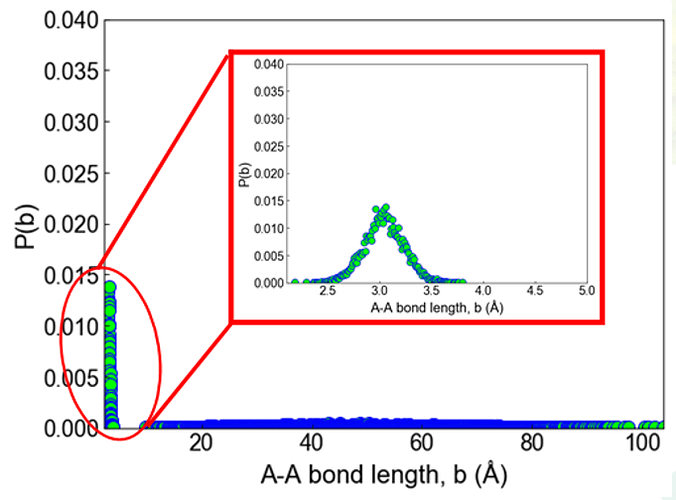
Recently, I noticed that lammps new version package named"lammps-patch_30Nov2020.tar.gz" added new bond and angle style, i.e., bond_style gaussian & angle**_style gaussian**. And I create a bead-spring chain to check the bond_style gaussian. I thought the final bond length distribution should be a single gaussian shape, but the result is not perfect, and the atoms are separated finally in system. The input file shown here:
############### Gaussian Potential Test ###############
units real
atom_style full
boundary p p p
bond_style gaussian
angle_style harmonic
dihedral_style harmonic
pair_style lj/gromacs 12.0 15.0
pair_modify mix arithmetic
read_data Beads.data
bond_coeff 1 300 1 0.0128 0.375 3.0 # New gaussian bond potential
angle_coeff 1 50.0 92.1
dihedral_coeff 1 0.111111 1 3
pair_coeff 1 1 0.0957 3.03315
neighbor 2.5 bin
neigh_modify delay 5
variable TI equal 1000 # Initial Temperature
variable TF equal 300 # Final Temperature
timestep 0.5
Gaussian_test.in (2.78 KB)
unwrap-1.dcd (391 KB)
Beads.data (777 Bytes)
All.lammpstrj (147 KB)