Hello all,
I am attempting to equilibrate, and eventually deform, a POPC lipid bilayer. However, I am running into a problem where all of my lipid molecules begin moving in the same direction during NPT. This movement is amplified when I attempt to deform the bilayer. This appears to be independent of lipid bilayer size and water amount.
I am using the 9Aug13 version of LAMMPS. I am currently running on 96 processors, but previously used 192 processors.
I do not understand where the motion could be coming from. Am I missing something obvious that would cause the molecules to begin moving?
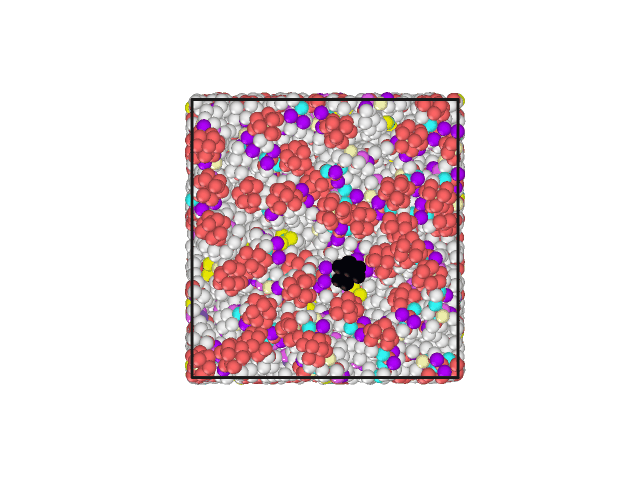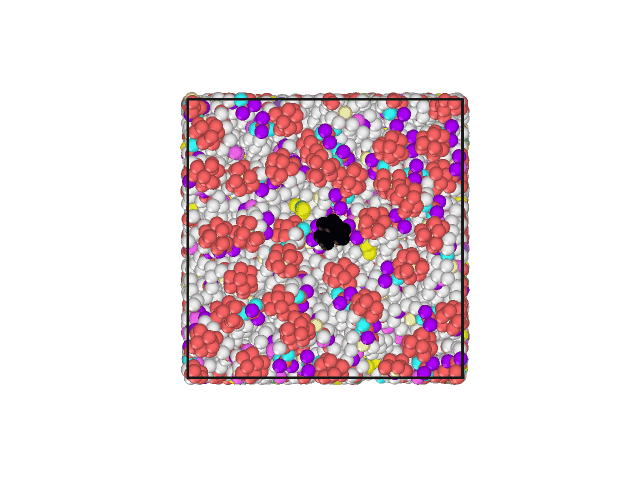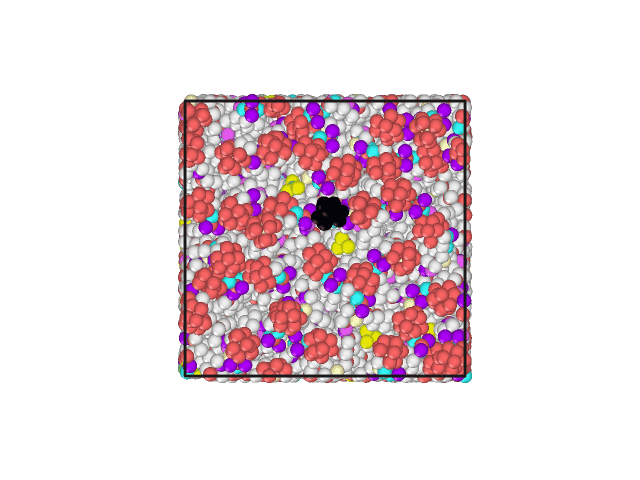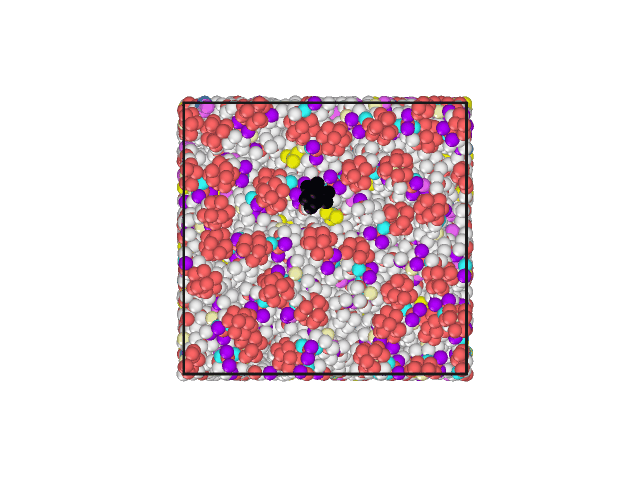
I have attached an animated gif displaying the top of the lipid bilayer. The water molecules are hidden. The lipid head group of one phospholipid has been changed to a different color to aid in visualization of movement.
Additional Information that may be useful:
Initial Structure:
I retrieved a 72 lipid (36/leaflet) POPC bilayer structure in pdb format from the site http://www.lobos.nih.gov/mbs/index.shtml which I converted to a LAMMPS data file using the charmm2lammps tool, a psf file produced in VMD, and the forcefield parameters for CHARMM36. I then changed the parameter values as directed in the LAMMPS documentation to use TIP3P water with long-range Coulombic solver and added more water. The structure has 36,858 atoms with the extra water.
Some Simulation Info:
I begin by initializing the velocities, moving the x and y dimensions of the box in to more closely fit the sides of the lipid bilayer, and ramping the temperature to 310 K while running NVT. I then change to NPT and run the simulation for the remainder of 3 ns (simulation is then restarted to run another 3 ns). I am using a .5 fs timestep. I have included portions of my input file:
---------- Define Simulation Parameters ------------------
units real
neigh_modify delay 0 every 1 check yes
atom_style full
bond_style harmonic
angle_style charmm
dihedral_style charmm
improper_style harmonic
pair_style lj/charmm/coul/long 8 10
pair_modify mix arithmetic
kspace_style pppm 1e-4
special_bonds charmm
---------- Define Thermo Style ------------------
thermo 10
thermo_style custom step elapsed cpu temp etotal pe ke press evdwl ecoul ebond eangle edihed eimp elong vol fmax fnorm
---------- NPT Portions of Simulation ------------------
#Uncoupled box with pressure = 1 atm and temperature = 310 K
fix 5 all npt temp 310 310 100 aniso 1 1 500
#Use “every” command without options during NPT to force kspace grid to reset
run 5700000 every 1000 “”
Results:
The Volume, Pressures, Energies all appear to become equilibrated (no net increase/decrease) during NPT. Additionally, the area per lipid appears to be acceptable. However, all of the lipids develop a motion in the same direction.
Other Comments:
Under advice of one of my committee members, I also ran a short NVE simulation starting after one of my equilibration runs (after 6 ns) which resulted in the potential, kinetic, and total energies having a slow increase.
I can include the complete input file or a data file (~8MB) if needed.
Thanks,
Michael