Dear all,
I am working in lammps. I used to fill a box with polymer chains of required density, using packmol and get lammps input file using VMD (topo tools) .
After minimization,
I used to get .dcd file as output. When loaded into VMD along with psf file (which was generated using VMD from intial pdb file), I find some unnecessary or unwanted , I dont know what it is , some kind of lines are forming here and there in VMD. Suddenly some atoms or part of polymer chain are hanging far away from my polymer chains. These polymer chains are attached to this atoms.
Could anyone explain some possible reason for this.
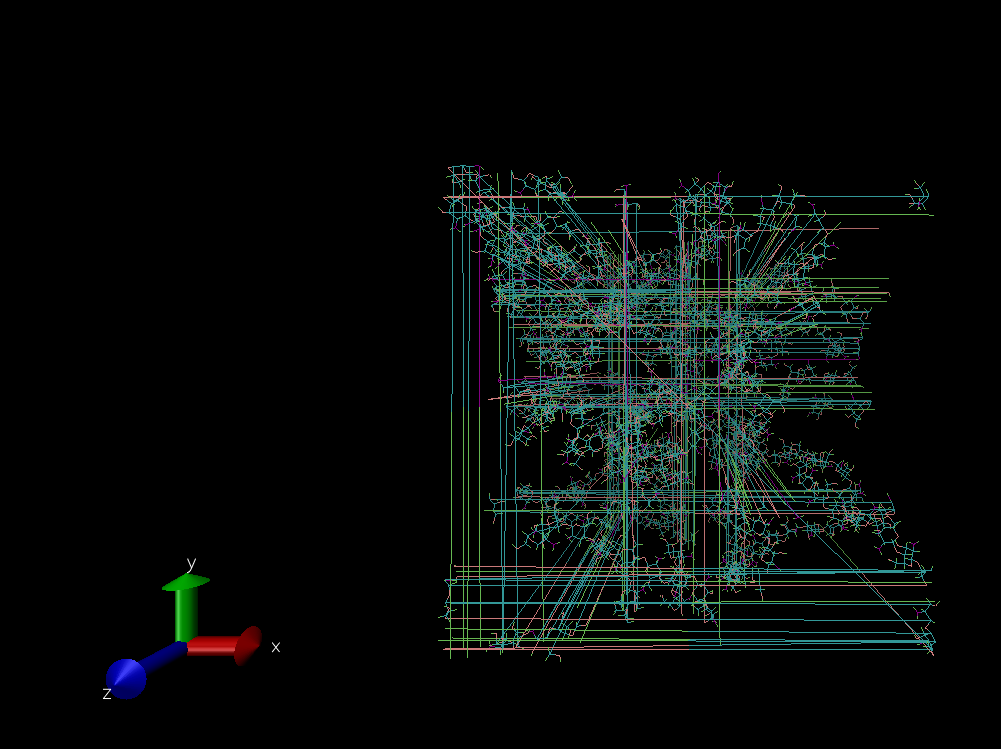
Thank you,
Dear all,
VMD questions are best asked on the vmd-l mailing list.
I am working in lammps. I used to fill a box with polymer chains of required density, using packmol and get lammps input file using VMD (topo tools) .
After minimization,
I used to get .dcd file as output. When loaded into VMD along with psf file (which was generated using VMD from intial pdb file), I find some unnecessary or unwanted , I dont know what it is , some kind of lines are forming here and there in VMD. Suddenly some atoms or part of polymer chain are hanging far away from my polymer chains. These polymer chains are attached to this atoms.
Could anyone explain some possible reason for this.
Wrapped coordinates, of course.
Axel
