Hello everybody
I’m working with rNEMD method for calculating the viscosity. The calculation is for a n-alkane system, in lammps-14May13, in three dimensions, with 512 molecules, and using real units.Part of the command that I am using is:
units real
atom_style full
boundary p p p
dimension 3
pair_style lj/cut/coul/long 10.0
pair_modify tail yes mix arithmetic
kspace_style pppm 1e-4
bond_style harmonic
angle_style harmonic
dihedral_style harmonic
improper_style none
read_data data.moleculanveeq
neighbor 2.0 bin
neigh_modify every 1 delay 0
timestep 1.0
fix 2 all nve
turn on Muller-Plathe driving force and equilibrate some more
velocity all scale 293.0
fix 4 all viscosity 200 x y 20
fix 5 all ave/spatial 50 200 10000 y center 2.0 vx units box
variable dVx equal f_5[11][3]-f_5[1][3]
thermo 10000
run 1000000
My problem is that the velocity slope dvx / dy is strongly non-linear. Based on the lammps manual, I should increase Nevery number in the fix viscosity command. So I increased it to 200, 500, 1000, but nothing improves the linearity of the velocity. I increased the Nevery, Nrepeat, and Nfreq of ave/spatial command too, be careful that the product of the first 2 numbers in the fix ave/spatial command will be smaller than the 3rd number, but nothing help.
I thought that my problem with the velocity could be caused by my kind of system or maybe, because I have something wrong in my code or is necessary use an additional command.
I would appreciate if you could give me some help, thank you very much
Ana
The fix ave/spatial command is simply monitoring what the velocity
profile is, it isn’t changing it. You don’t say how the profile is non-linear.
There are many reasons it could be. Not long enough equilibration, entanglement
of chains, etc. If you post a plot of the velocity profile, maybe someone
will have ideas.
You are also using a version of LAMMPS that is 1.5 years old. I suggest
you try the current version.
Steve
Thanks for try to help me
In my simulations I use 1 fs of time step. The system was equilibrated 2ns in NVT ensemble and 1ns in NVE ensemble. In addition, using fix viscosity command the total time of simulation was 3ns. One plot of the velocity profile for the system that I showed in the last message is the following:
Spatial-averaged data for fix 5 and group all
Timestep Number-of-bins
Bin Coord Ncount vx
4000000 22
1 1.4372 271.2 -0.000236024
2 3.4372 361.5 -0.00024852
3 5.4372 345.05 -0.000317888
4 7.4372 367.05 -6.24965e-05
5 9.4372 362.9 -0.000125705
6 11.4372 351.05 -0.00017553
7 13.4372 366.05 -8.6322e-05
8 15.4372 351.6 -0.000331148
9 17.4372 354.7 -3.50788e-05
10 19.4372 363 8.30432e-06
11 21.4372 355.35 -0.000195202
12 23.4372 358.85 -3.13579e-07
13 25.4372 364.2 -4.43861e-05
14 27.4372 349.4 5.61896e-05
15 29.4372 364.6 -7.7006e-05
16 31.4372 364.3 -0.000174875
17 33.4372 342.2 -0.000132875
18 35.4372 365.5 -0.000211639
19 37.4372 352.85 -0.00012789
20 39.4372 359.25 -0.000237023
21 41.4372 352.6 -0.00015073
22 43.4372 256.8 -0.000214508
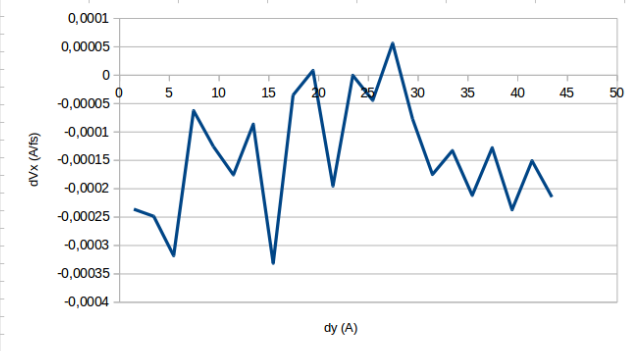
The plot is clearly non-linear. I have been thinking in use a shake command in the equilibration part (not when I use fix viscosity because It is a mistake) or remove the angle_style and the dihedral_style, but I don’t know if it is a good idea or help to solve my problem.
Ana
Those are tiny velocities. I’m not convinced anything
is actually moving much. Have you visualized your system?
You should see a clear V-shaped velocity profiles just
from animating it. Like the examples provided with LAMMPS.
Steve

