Hi there,
I have been working on a code which retrieves XAS documents from the materials project database then uses the material id of the retrieved compounds to retrieve their structure documents. A copy of these sections of the code are provided below:
MP_API_KEY =
from emmet.core.xas import Edge, Type
from mp_api.client import MPRester
req_fields = ["material_id", "structure", "composition", "symmetry", "nsites", "sites", "elements", "spectrum"]
with MPRester(MP_API_KEY,use_document_model = False, monty_decode = False) as mpr:
zn_docs = mpr.materials.xas.search(edge=Edge.K,
spectrum_type=Type.XANES,
absorbing_element="Zn",
fields = req_fields)
Zn_struct_doc = []
for i in range(len(zn_docs)):
C = mpr.get_structure_by_material_id(Zn_mat_id[i])
Zn_struct_doc.append(C)
What I am finding is that while the list in the Zn structure docs is of the same size as zn_docs, some of the elements are empty. Thus:
- Does this mean that not all the materials in the zn-docs list have computed structures?
- Is there a problem with the endpoint?
Thanks for reaching out. Do you have an example mp-id that results in an empty element? Also, where is Zn_mat_id defined?
Hello! Thank you for the quick response!
The Zn_mat_id is defined as follows:
Zn_mat_id = [[] for i in range(len(zn_docs))]
for i in range(len(zn_docs)):
Zn_mat_id[i] = zn_docs[i]["material_id"]
The first 10 entries in the Zn_mat_id list are as follows:
mp-1041683
mp-35033
mvc-4383
mp-36165
mp-1043579
mvc-5133
mp-1042240
mp-9571
mp-1045659
mp-979012
And the corresponding structure docs are as follows:
Run the mp-id from the XAS search through mpr.get_material_id_from_task_id() (see here) and use the resulting mp-id in mpr.get_structure_by_material_id(). That’ll get you the correct mp-id for the ones with the mvc- prefix.
Also, for the benefit of other users stumbling upon this thread, the more pythonic way of writing the loop is
Zn_struct_doc = [
mpr.get_structure_by_material_id(doc["material_id"])
for doc in zn_docs
]
@munrojm is the structure in xas.search any different from the one returned through get_structure_by_material_id()?
Thank you! I have implemented the new line of code as follows:
Zn_material_ids = [[] for i in range(len(zn_docs))]
Zn_material_ids_2 = [[] for i in range(len(zn_docs))]
for i in range(len(zn_docs)):
Zn_material_ids[i] = zn_docs[i]["material_id"]
for i in range(len(zn_docs)):
with MPRester(MP_API_KEY) as mpr:
Zn_material_ids_2[i] = mpr.get_material_id_from_task_id(Zn_material_ids[i])
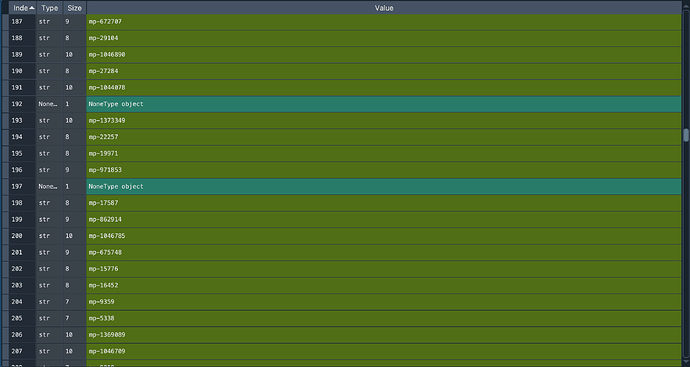
This does find the canonical mp ids for all the documents, but I occasionally find the following in the list (see entry 192 and 197):
I am assuming that this is because the task id can’t be found?
Yes, that could be the case. What are the task-ids for these two entries?
Hi,
The task id for entry 192 is mp-1703 and for 197 it is mp-980062.
It should be the same structure.
– Jason
Hey Matt,
My original XAS rester query is parsed as follows (there seems to a fault with the serializer):
MP_API_KEY =
from emmet.core.xas import Edge, Type
from mp_api.client import MPRester
req_fields = ["material_id", "task_ids", "structure", "composition", "symmetry", "nsites", "sites", "elements", "spectrum"]
with MPRester(MP_API_KEY,use_document_model = False, monty_decode = False) as mpr:
zn_docs = mpr.materials.xas.search(edge=Edge.K,
spectrum_type=Type.XANES,
absorbing_element="Zn")
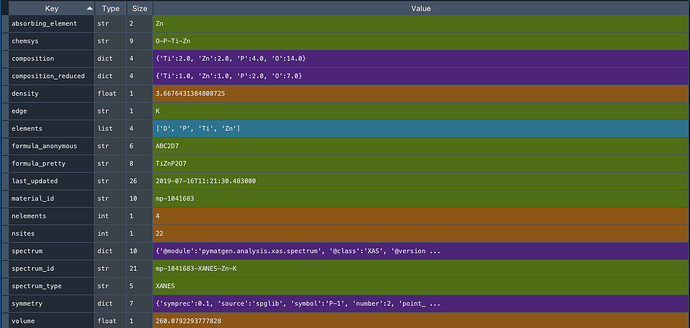
The output from zn_docs[0] is as follows:
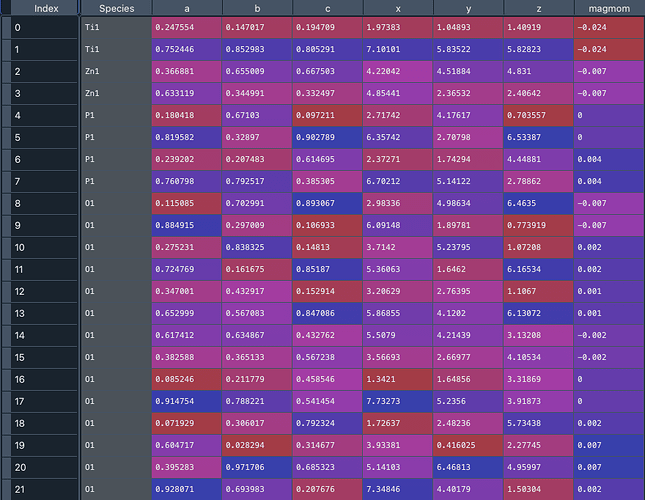
What I would like is this (for the same material_id but from the structure rester):
This allows me to compute bond lengths, coordination numbers, etc from the pymatgen structure. It could be that there is a more direct way to calculate this information, but my current workflow is to extract relevant structural information from the structures (in this case Zinc compounds) whose XAS spectra are computed.
Thanks.