Dear Alex,
I want to know how to I enter the command to get the following two image?(Average scattering rates as a function of temperature and charge carrier
concentration)
Looking forward to your answer!
Dear Alex,
I want to know how to I enter the command to get the following two image?(Average scattering rates as a function of temperature and charge carrier
concentration)
Hi Jincheng,
This isn’t available straight from AMSET as far as I know, but I’ve written a code, ThermoPlotter, which has this as a command-line tool. The command is tp plot avg-rates <mesh-file(s)>.
Hope this helps!
Kieran
Thank you very much for your help. I have one more question for you. Whether the crystal structure of the AMSET input is specific to the requirements, such as the need to use orthogonal unit cells. Looking forward to your reply!
The main requirements for the crystal structure:
Thank you, sir, for everything you do for this, and I will continue to follow you!
Dear Alex,
The elastic matrix of the strongly anisotropic material I calculated contains negative numbers, and the settings.yaml file is set as follows. I’m not sure if it’s the right thing.
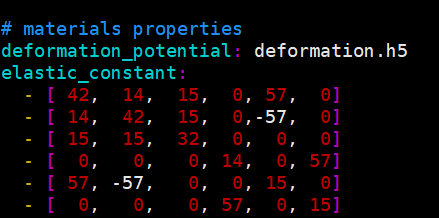
You can use this script to extract the elastic constants. Simply download the script, make it executable and run ./elastic OUTCAR in the elastic constant calculation folder.
However, negative numbers are not always bad. When the appear in the off diagonal elements that can occur when you don’t have an orthogonal lattice.
Dear Alex,
Thank you very much for your reply!
Best,
Jincheng
Dear Kieran,
ThermoPlotter does not support “.dat”?
I tried to use plot avg-rates, but it only supports figure file.
Thanks!
Hi Euicheol,
Do you mean it doesn’t support writing the data to “.dat”? This can’t be done at the command line at present, but it could be done with a short python script. The data can be calculated from the mesh.h5 file using data = tp.data.load.amset_mesh(<mesh_file>, 'weighted_rates'), which you can then save to .dat using your preferred method.
data['weighted_rates'] will be three dimensional (scattering type, temperature, doping), so you may need several files if you save all of it. You can find out which scattering type corresponds to which line by checking data['stype'], and likewise for the other dependent variables. Other useful information can be found in data['meta'].
Hope this helps,
Kieran
I try to your comments
Really appreciate!
Euicheol
Dear Alex,
When collecting deformation potential files, I made sure that the same K-mesh was used for each file, and the following error occurred.I’m not sure where this 40 came from.
Please tell me how to solve it. I also encountered the same problem.