I am a little bit confused about the results of my grain segmentation and polyhedral template matching. I am simulating a single crystal of lithium, and holding it at 300K using an NVT thermostat for 10 nanoseconds. The Voronoi analysis of the final state after minimization returns the majority of the atoms having 14 coordination, as expected for BCC. However, the grain segmentation identifies the grains as majority either HCP or FCC. Is this a reflection of my simulation or of the experimental nature of grain segmentation currently?
Hi,
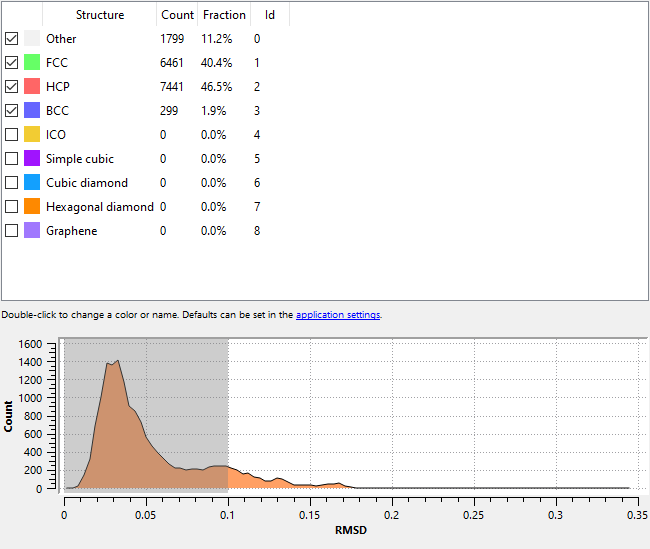
Could you please describe (or show us) how the output of the Polyhedral Template Matching (PTM) function looks like in your case? It’s the PTM that marks atoms as either HCP, FCC or BCC, not the grain segmentation algorithm.
Yes, the PTM outputs the majority of atoms as having FCC or HCP.
Bond-based CNA gives slightly more expected results, with about 40% being identified as BCC and the rest classified as ‘other’.
I cannot really comment on your simulation, because I haven’t seen it and don’t know the details. But generally I would trust PTM more than CNA to determine the more likely crystal structure. PTM has been designed with a well-defined metric in mind measuring the distances of the actual structure from all ideal candidate structures. If PTM says it’s FCC/HCP and not BCC, then it really means the structure in your simulation is more FCC/HCP-like, i.e. more close-packed than you expected.
Using bond-based CNA for this purpose is dangerous, because the bonds you create before the CNA analysis introduces a bias. If you give an atom 14 neighbour bonds, then bond-based CNA is coerced into considering only BCC as a possible candidate structure. FCC and HCP are not even considered, because they would require 12 bonds.