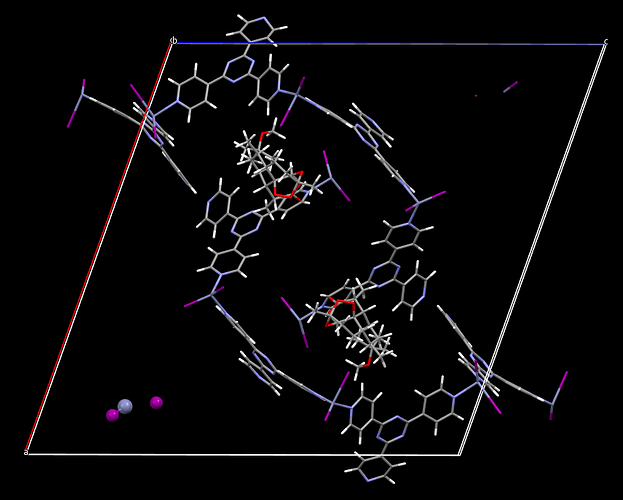
I am attempting to run an optimisation of a MOF structure in GULP using the UFF4MOF force field, however, I am having problems with bonds that cross the periodic boundary. The optimisation seems to be running okay until I check the structure, and I can see that some of the metal-halide nodes (those which are across the periodic boundary) separate from the organic ligands. See bottom left of image:
I can only assume this is because the bonding isn’t being described correctly, but I don’t know how to fix this. I have followed advice from a previous post (Wrong bonding in output cif file - #4 by Michal_Husak) to get GULP to describe the bonding and also the types of bonds in a dump file, which I believe is correct for this structure.
For some reason I cannot upload the .cif of the extended MOF framework I am trying to model, but the input I used for the calculation is below.
Any help is greatly appreciated!
opti conp molmec
title
XAZPEW bond test
end
cell
34.69368894 14.85721046 34.56270034 90.0 108.680920 90.0
fractional
Zn1 core 0.1713 -0.2211 0.0162
I1 core 0.1204 -0.2463 -0.0542
I1 core 0.235 -0.3181 0.0456
Zn1 core -0.1107 0.0133 0.195
I1 core -0.101 0.1682 0.1677
I1 core -0.1213 -0.0171 0.263
N1 core 0.1479 0.4773 -0.1153
N1 core 0.1881 -0.0855 0.0207
N1 core 0.3435 0.4483 0.1498
N1 core 0.1891 0.245 -0.0043
N1 core 0.2437 0.2298 0.0575
N1 core 0.2296 0.371 0.0241
N1 core 0.1413 -0.2303 0.0601
N1 core -0.0624 -0.0639 0.1903
N1 core 0.114 -0.5605 0.2866
N1 core 0.0473 -0.2155 0.1462
N1 core 0.0433 -0.2972 0.2037
N1 core 0.0941 -0.3333 0.1743
C1 core 0.1481 0.3868 -0.1152
H1 core 0.1361 0.3531 -0.1444
C1 core 0.1634 0.3375 -0.0797
H1 core 0.1633 0.2648 -0.081
C1 core 0.1798 0.3831 -0.0429
C1 core 0.1789 0.4771 -0.0428
H1 core 0.1913 0.5152 -0.0148
C1 core 0.1623 0.5217 -0.0795
H1 core 0.1602 0.5946 -0.0808
C1 core 0.2156 -0.05 0.0541
H1 core 0.2309 -0.0972 0.0781
C1 core 0.2244 0.0411 0.058
H1 core 0.2463 0.0669 0.0857
C1 core 0.2042 0.0983 0.0259
C1 core 0.1761 0.0613 -0.0089
H1 core 0.1604 0.1038 -0.0345
C1 core 0.1689 -0.0301 -0.0104
H1 core 0.1473 -0.0623 -0.0367
C1 core 0.3163 0.4964 0.1204
H1 core 0.32 0.569 0.1223
C1 core 0.2857 0.4559 0.0896
H1 core 0.2643 0.4966 0.0665
C1 core 0.2827 0.3622 0.0887
C1 core 0.3106 0.3125 0.1191
H1 core 0.309 0.2399 0.12
C1 core 0.3405 0.3578 0.1492
H1 core 0.3629 0.3212 0.1732
C1 core 0.1999 0.331 -0.0054
C1 core 0.2126 0.1962 0.027
C1 core 0.2503 0.3183 0.0554
C1 core 0.1029 -0.1997 0.0522
H1 core 0.0892 -0.1698 0.0223
C1 core 0.0818 -0.2082 0.0799
H1 core 0.0511 -0.1828 0.0728
C1 core 0.1005 -0.2518 0.1169
C1 core 0.1402 -0.2828 0.1251
H1 core 0.1555 -0.3167 0.1536
C1 core 0.1596 -0.2703 0.0963
H1 core 0.1904 -0.2938 0.1011
C1 core -0.0403 -0.1164 0.2211
H1 core -0.0486 -0.1142 0.2488
C1 core -0.0093 -0.1706 0.2175
H1 core 0.008 -0.2127 0.2426
C1 core -0.0008 -0.1711 0.1807
C1 core -0.0236 -0.1164 0.1488
H1 core -0.0179 -0.116 0.1198
C1 core -0.0541 -0.0638 0.1548
H1 core -0.0724 -0.0191 0.1312
C1 core 0.1404 -0.5177 0.2716
H1 core 0.1718 -0.5393 0.2831
C1 core 0.1283 -0.4496 0.243
H1 core 0.1507 -0.4167 0.2322
C1 core 0.0878 -0.4225 0.2304
C1 core 0.0605 -0.4656 0.2464
H1 core 0.029 -0.4445 0.2374
C1 core 0.0746 -0.535 0.2739
H1 core 0.0548 -0.5727 0.2867
C1 core 0.0792 -0.2674 0.1472
C1 core 0.0315 -0.2308 0.1763
C1 core 0.0744 -0.3474 0.2013
Zn1 core 0.1349 -0.6624 0.3295
I1 core 0.0787 -0.7726 0.3257
I1 core 0.2046 -0.7086 0.3244
Zn1 core 0.1342 0.5484 -0.1699
I1 core 0.076 0.6546 -0.1756
Zn1 core 0.6731 -0.3979 0.5225
Zn1 core 0.3881 -0.6341 0.6984
I1 core 0.3744 -0.6076 0.765
I1 core 0.4007 -0.7878 0.6712
N1 core 0.1508 -0.5913 0.3839
N1 core 0.1887 -0.0324 0.5258
N1 core 0.3438 -0.5676 0.6521
N1 core 0.1956 -0.3584 0.4943
N1 core 0.2488 -0.3447 0.5572
N1 core 0.2362 -0.4844 0.5224
N1 core 0.4372 -0.5573 0.6951
N1 core 0.6135 -0.0541 0.7872
N1 core 0.6393 -0.3906 0.5631
N1 core 0.5425 -0.3218 0.7077
N1 core 0.5913 -0.284 0.676
N1 core 0.5466 -0.4067 0.651
C1 core 0.1502 -0.5007 0.384
H1 core 0.1361 -0.4673 0.355
C1 core 0.1668 -0.4512 0.4191
H1 core 0.1659 -0.3785 0.4179
C1 core 0.1858 -0.4963 0.4556
C1 core 0.1857 -0.5903 0.4557
H1 core 0.1999 -0.6284 0.4833
C1 core 0.1676 -0.6354 0.4194
H1 core 0.1663 -0.7083 0.4184
C1 core 0.2163 -0.0691 0.5588
H1 core 0.2306 -0.0237 0.5838
C1 core 0.2265 -0.1594 0.5607
H1 core 0.2487 -0.1863 0.5879
C1 core 0.2077 -0.2142 0.5272
C1 core 0.1789 -0.1761 0.4933
H1 core 0.1636 -0.2172 0.467
C1 core 0.1701 -0.0856 0.4936
H1 core 0.1483 -0.0523 0.4679
C1 core 0.3192 -0.6138 0.6202
H1 core 0.3234 -0.6862 0.6209
C1 core 0.2905 -0.5716 0.5881
H1 core 0.2712 -0.6109 0.5631
C1 core 0.2867 -0.4781 0.5887
C1 core 0.3119 -0.4304 0.6219
H1 core 0.3098 -0.358 0.6238
C1 core 0.3399 -0.4773 0.653
H1 core 0.3601 -0.4422 0.6792
C1 core 0.2066 -0.444 0.4928
C1 core 0.2179 -0.3107 0.5266
C1 core 0.2559 -0.4328 0.5544
C1 core 0.4567 -0.499 0.7247
H1 core 0.4463 -0.4971 0.7511
C1 core 0.4877 -0.4444 0.7213
H1 core 0.503 -0.3978 0.7454
C1 core 0.4989 -0.4497 0.686
C1 core 0.4788 -0.5106 0.6555
H1 core 0.4868 -0.5158 0.6278
C1 core 0.4482 -0.5632 0.6612
H1 core 0.432 -0.6126 0.6388
C1 core 0.6388 -0.0935 0.7699
H1 core 0.6696 -0.067 0.7791
C1 core 0.6264 -0.1629 0.7421
H1 core 0.6473 -0.1934 0.7285
C1 core 0.5867 -0.1946 0.7325
C1 core 0.5604 -0.1543 0.7506
H1 core 0.5295 -0.1783 0.7436
C1 core 0.5747 -0.0836 0.7773
H1 core 0.5556 -0.048 0.7916
C1 core 0.6153 -0.4566 0.5694
H1 core 0.614 -0.5189 0.5527
C1 core 0.5938 -0.4471 0.5968
H1 core 0.5751 -0.5017 0.6013
C1 core 0.5974 -0.3672 0.6188
C1 core 0.622 -0.2991 0.6119
H1 core 0.6252 -0.2365 0.6285
C1 core 0.6422 -0.3129 0.584
H1 core 0.6624 -0.2628 0.5783
C1 core 0.5309 -0.3899 0.6811
C1 core 0.5727 -0.2707 0.7039
C1 core 0.577 -0.3525 0.6499
I1 core 0.2046 0.5953 -0.1713
I1 core 0.6302 -0.3596 0.4502
I1 core 0.7357 -0.2987 0.5583
O3 core 0.3698 0.0451 0.3651
O3 core 0.3765 0.0118 0.4071
O3 core 0.3137 0.0581 0.4104
O3 core 0.3431 0.1978 0.4146
O2 core 0.3804 0.322 0.4271
C1 core 0.2363 0.059 0.2566
H1 core 0.2517 0.0314 0.2361
H1 core 0.2126 0.1048 0.2386
H1 core 0.2207 0.0042 0.2667
C1 core 0.2662 0.1105 0.2922
H1 core 0.2489 0.1338 0.3119
C1 core 0.3013 0.0501 0.3184
H1 core 0.3177 0.0236 0.2984
C1 core 0.334 0.1042 0.3513
C1 core 0.3492 0.1873 0.3341
H1 core 0.3638 0.1616 0.3124
C1 core 0.3137 0.2479 0.31
H1 core 0.3255 0.3034 0.2963
H1 core 0.2989 0.2788 0.3305
C1 core 0.2825 0.1943 0.2764
H1 core 0.2568 0.238 0.2609
H1 core 0.2962 0.1739 0.2532
C1 core 0.2851 -0.0302 0.3371
H1 core 0.2718 -0.0797 0.3131
H1 core 0.2601 -0.0069 0.3475
C1 core 0.316 -0.0798 0.3728
H1 core 0.2995 -0.1316 0.3836
H1 core 0.3391 -0.1136 0.3627
C1 core 0.3385 -0.0197 0.4095
C1 core 0.3489 -0.0652 0.4508
H1 core 0.3649 -0.018 0.4748
H1 core 0.3684 -0.1236 0.4519
H1 core 0.3209 -0.0873 0.4559
C1 core 0.3184 0.131 0.3865
H1 core 0.2885 0.1623 0.3742
C2 core 0.3687 0.257 0.405
C1 core 0.3826 0.234 0.3689
H1 core 0.4061 0.1825 0.3812
C1 core 0.4025 0.3146 0.3553
H1 core 0.3819 0.372 0.3467
H1 core 0.4117 0.297 0.3289
H1 core 0.4295 0.336 0.38
O3 core 0.2189 -0.3888 0.2826
C1 core 0.2605 -0.4174 0.288
C1 core 0.2795 -0.4588 0.3306
H1 core 0.2815 -0.4099 0.3548
H1 core 0.3103 -0.4827 0.3343
H1 core 0.2613 -0.5161 0.3345
C1 core 0.2128 -0.3134 0.3058
H1 core 0.1802 -0.3096 0.3007
H1 core 0.2224 -0.2501 0.2954
H1 core 0.2284 -0.3213 0.3387
C1 core 0.2556 -0.4891 0.255
H1 core 0.2377 -0.5458 0.2602
H1 core 0.2852 -0.5141 0.2549
H1 core 0.2398 -0.4606 0.2248
C1 core 0.2862 -0.3386 0.2811
H1 core 0.2706 -0.3054 0.252
H1 core 0.3156 -0.3635 0.2801
H1 core 0.2921 -0.2885 0.3055
species
Zn1 core Zn3f2
H1 core H_
C1 core C_R
C2 core C_2
C3 core C_3
N1 core N_R
O1 core O_R
O2 core O_2
O3 core O_3
I1 core I_
space
C 2
library /mainfs/home/$USER/programfiles_GULP/gulp-6.2/Libraries/uff4mof.lib
bondtype C1 C1 resonant
bondtype C1 N1 resonant
bondtype C2 C2 double
bondtype C2 O2 double
bondtype C3 O3 single
bondtype O3 O3 single
bondtype Zn1 I1 single
bondtype Zn1 N1 single
dump connect bondtest.res
dump every 10 bondtest_opt.res
output cif bondtest_opt