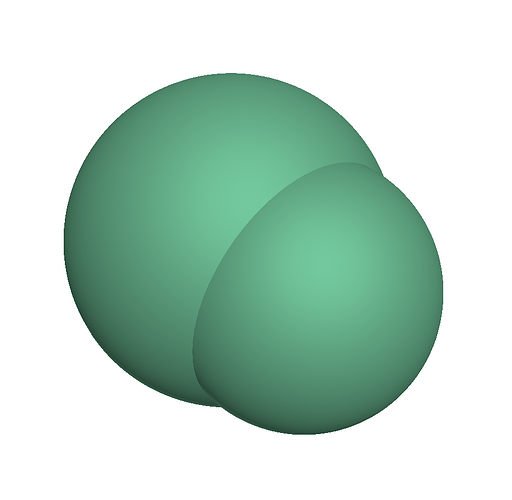Dear Advisers in this Society,
I am simulating the mechanics of rigid clumps generated using a molecule template.
To verify the properties of clumps, I am using a minimum working example with the simplest clumps, as shown below.
The clumps are generated into the simulation domain using the fix pour command.
From my understanding of the LAMMPS manual(molecule command — LAMMPS documentation), the properties of a clump should be predefined when the clump consists of overlapping atoms.
“For mass, com, and inertia, the default is for LAMMPS to calculate this quantity itself if needed, assuming the molecules consist of a set of point particles or finite-size particles (with a non-zero diameter) that do not overlap. If finite-size particles in the molecule do overlap, LAMMPS will not account for the overlap effects when calculating any of these 3 quantities, so you should pre-compute them yourself and list the values in the file.”
Therefore, I calculated the relevant properties (mass, center of mass, and moment of inertia) using Monte Carlo integration and provided them in the molecule template file (ms_v2.data).
# Monte Carlo Molecule for Clumps
# header section:
2 atoms
1418.7035714997178 mass
2.5339358941207264 5.410052941769989 6.794372462111523 com
157.9662266218661 155.89781209308427 126.63528407308945 1.845998558936373 -5.580695808688286 9.970510762159702 inertia
# body section:
Coords
1 2.496793269872739 5.474982340911671 6.569003413784817
2 2.5444160863821272 5.391228984360708 6.857014284485096
Diameters
1 0.7525078827946253
2 0.9524418398136981
Types
1 1
2 1
After checking the properties using compute rigid/local and dump local, I found that LAMMPS does not use the values from the molecule template. Instead, it sums the properties of the individual atoms, counting the overlapping portions twice.
ITEM: NUMBER OF ENTRIES
10
ITEM: BOX BOUNDS ff ff ff
-1.0000000000000000e+01 1.0000000000000000e+01
-1.0000000000000000e+01 1.0000000000000000e+01
0.0000000000000000e+00 5.0000000000000000e+01
ITEM: ENTRIES c_get_rigid_properties[1] c_get_rigid_properties[2] c_get_rigid_properties[3]
6 3 0.675508
4 2 0.675508
12 6 0.675508
14 7 0.675508
16 8 0.675508
20 10 0.675508
2 1 0.675508
10 5 0.675508
18 9 0.675508
8 4 0.675508
LAMMPS gives the mass of the clump as 0.675508, which corresponds to the sum of the volumes of each atom (0.223 + 0.425), assuming the atomic density is 1. However, the mass of the molecule is provided as “1418.7035714997178 mass” in the template
My question is: In this cases, Are the properties output through compute rigid/local actually used for the calculation of clump mechanics, or are they only for compute and dump purposes?
Additionally, I found in the fix/rigid documentation(fix rigid command — LAMMPS documentation) that for overlapping clumps, properties should be provided using the infile keyword with an external file:
"The infile keyword allows a file of rigid body attributes to be read in from a file, rather than having LAMMPS compute them. There are 5 such attributes: total mass, center-of-mass position, 6 moments of inertia, center-of-mass velocity, and 3 image flags for the center-of-mass position.
For rigid bodies consisting of point particles or non-overlapping finite-size particles, LAMMPS can compute these values accurately.
However, for rigid bodies consisting of overlapping finite-size particles, LAMMPS will ignore the overlaps when computing these attributes, leading to incorrect dynamics. The error depends on the amount of overlap. To avoid this issue, the values should be precomputed (e.g., using Monte Carlo integration)."
However, I believe that the infile cannot be generated before creating clumps when using fix pour, since the clump structure is determined dynamically.
Could you please clarify if there is a way to correctly apply predefined clump properties in this case, or have I misread the manual?
Thank you for your time and insights.
Best regards,
MWE
# 1. setup variables and calculate critical time steps
units si
molecule clumps_01 ms_v2.data #scale 1.0
timestep 1E-5
# 2. setup simulation environments
atom_style hybrid sphere molecular
newton on
boundary f f f
dimension 3
neighbor 1.0 bin
comm_modify vel yes
neigh_modify delay 0 every 10 check yes once no cluster no exclude molecule/intra all page 10000000 one 20000
region domain_3D block -10.0 10.0 -10.0 10.0 0.0 50.0
create_box 2 domain_3D
# 4. setup walls
region bottom_plate plane 0.0 0.0 0.0 0.0 0.0 1.0 side in
fix bottom all wall/gran/region hertz/history 1E9 1E6 1E6 1E3 0.5 1 region bottom_plate
# 5. Read clumps and setup pairs, EQ of motions
region gen_area block -5.0 5.0 -5.0 5.0 20.0 40.0
fix make_clumps_1 all rigid/small molecule mol clumps_01 gravity grav_acc
compute adjust_DOF all temp/sphere
thermo_modify temp adjust_DOF
pair_style gran/hertz/history 1E9 1E6 1E6 1E3 0.5 1
pair_coeff * *
group temp_rigid empty
#fix grav_acc all gravity 9.81 vector 0.0 0.0 -1.0
fix grav_acc temp_rigid gravity 9.81 vector 0.0 0.0 -1.0
fix viscous_damping all viscous 0.0001
# 6. Setup dump
#shell if [ -d "post_1" ]; then rm -rf post_1; fi # make directory for post
#shell mkdir post_1
#dump dump_atoms all vtk 1000 post_1/atoms*.vtk fx fy fz xu yu id type radius diameter mol x y z vx vy vz
fix pour_clumps1 all pour 10 0 4767548 region gen_area mol clumps_01 molfrac 1.0 rigid make_clumps_1
run 100000
compute get_rigid_properties all rigid/local make_clumps_1 id mol mass
dump dump_rigid_properties all local 1000 temp.dump c_get_rigid_properties[1] c_get_rigid_properties[2] c_get_rigid_properties[3]
run 100000
ms_v2.data (494 Bytes)
in.MWE250312 (1.9 KB)