Axel,
Thanks for your response. I followed your suggestion. I used pair_style hybrid and considered lj interactions for atoms close to surface and bonded interactions for the rest, and the problem is solved. There are just few things
Dear Lammps users,
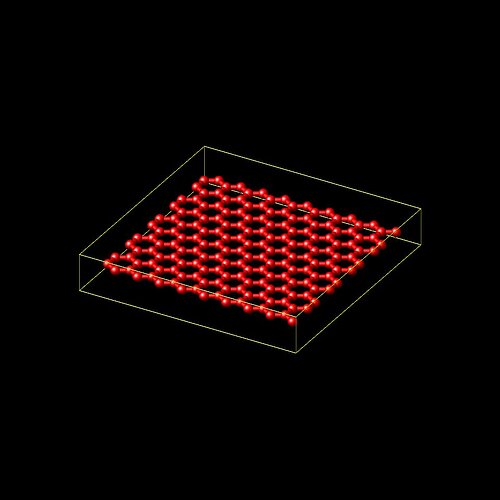
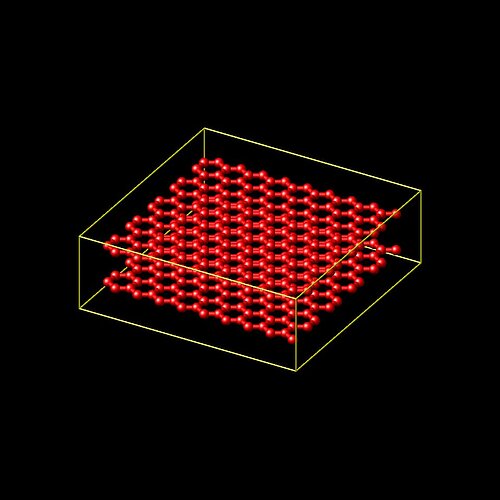
I want to perform the compression test on a system which is consist of two layers are on top of each other but I want the layers to still be treated as two separate layers, without interaction between the. I mean, consider having two sheets of paper on top of each other. For this case, generating the simulation system is not as easy as just making two layers on top of each other.
i disagree. it is. this is how you would model graphite, for example. and there have been plenty of studies of multiple graphene layers on top of each other. there are even examples for that bundled with LAMMPS which have some specific inter-layer potential overlays to correct the otherwise not perfectly accurate inter-layer interactions of commonly used potentials. but the simplest model is bonded interactions (bonds, angles and dihedrals) within the layer and for interactions between pairs farther than 4 atoms apart and between layers regular lj/cut interactions.
By this paragraph I did not mean it is not possible, I said it is not as easy as duplicating system. This is something that might be clear for some people, but not for me, and I was happy realizing it  .
.
We can not just replicate the first layer to generate the the second one. Because by replication bonds and other topology interactions are created between pairs of new atoms as well as between old and new atoms.
that is not correct. the replicated system will be a replica of the original system and nothing more. the interactions between the layers are purely non-bonded interactions, and those would be physical.
This statement is exactly from the manual, I am not sure why you say it is not correct. I mean if I do not change the interactions between atoms, which I was not aware of, and I just create a system and duplicate it, there will be bonds between the new and old atoms. And you were right, by using different interaction for bulk and surface atoms, the problem is solved.
Thanks again for your time and help.
Best,
Sepideh
Lammps will not distinguish between the first and second layer.
LAMMPS will compute the forces as described in the model you define.
Using “lattice” and “create atom” command for each layer, will generate the system such that the two layers are bonded (glued) to each other, while we want each layer to move freely.
well, if you have the same interaction between the atoms everywhere, how should the physics distinguish them? it will just compute forces between them.
I selected the atoms close to top boundary of the layer on the bottom , atoms close to the bottom of the top layer on the top, and turned off the pairwise interaction between those groups. But this time when I apply compression, the selected atoms do not interact with each other and atoms in those groups enter each other, because they do not repeal each other any more.
of course. if you have no interactions, there cannot be any repulsion. if you want to layers to “see” each other, they need some potential between them.
the simplest approach is to have different interactions between the layers than within the layer. see my comment from above.
axel.