Dear LAMMPS representatives:
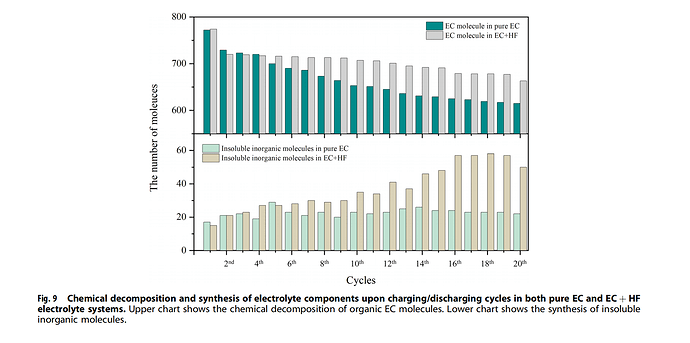
Are there any commands in LAMMPS can count the total number of a species from the output file? I want to get a result as the following picture, to count EC molecules number in each step.
Or do I need write a code myself to determine a molecule type from the distances of each atoms?
Thank you.
Hi,
It is very unclear what you want. You want to count a number of species from an output (what output? a dump file?) for each step? and also you want to count molecule within a certain distance from some other atoms?
Very confusing.
Yes, LAMMPS has many commands that you can include in the input, not the output to measure the number of species, see the ‘count’ variable for instance. For species within a distance from another species, see either the rdf or the coord/atom computes. For measuring something from the output, whatever the output is, probably post-processing tools would be best.
Simon
ps: Adding graph without context bring little information, particularly if the graph contains acronyme.
Sorry I didn’t make it clearly. Yes, I want to count the number of ethylene carbonate (EC) from the dump file, as this picture shows.
I wonder if the molecule ID or any other settings in the input file could help me find each intact ethylene carbonate molecule in the output file.
As the picture below, it’s hard to tell how many ethylene carbonate molecules() are there from the image generated by OVITO.
It may be possible to that on-the-fly using LAMMPS, but I am not sure.
It is definitely possible to do that using postprocessing. The postprocessing tool I like is MDAnalysis/python, which allow you to read a lammps dump file.
Thank you very much, don’t know this before, I will try find solutions there
I think that if you plan on doing this from a dump file you already collected, there is no other way other than postprocessing. But if you are willing to re-run the simulation, I have an idea that came to mind to get it on the flight (but you need to check on the manual if indeed this will work).
If your EC molecules contain atoms that are of a specific type, and such atom types only occur in the EC molecules, maybe you can set up an atom-style variable command which computes, for example, the mass (see variable command — LAMMPS documentation for more details). Then, you can couple this to a compute reduce sum to output the information at different time steps (see compute reduce command — LAMMPS documentation for more). And ofc you would need to set up a group-ID for this compute command that only encapsulates the atom types of the atoms in the EC molecules. You can plug the ID of the compute reduce in the thermo_style command to get instant values of total mass of EC molecules at a given time instant (see thermo_style command — LAMMPS documentation for more). You can then figure out how many molecules you actually have at different time steps by reasoning: “if I have this total mass of EC molecules, how many EC molecules do I have in my system?”
Thank you, but those atom types (C, H, O) in EC molecules are shared with other molecules in SEI, in this case it seems I need postprocessing to do so.
You can change your setup to have different atom types for the same atoms that are in different kinds of molecules and use the same potential parameter settings.
However, also this sounds a lot like you are using ReaxFF (this is important information if you want good advice) and in that case, there is fix reaxff/species which can classify molecules by processing the bond order information. You can also retrieve the number of molecules and the number of distinct species from that fix as well as a per-atom property that can be used instead of molecule IDs.
For non-reactive simulations, the categorization of atoms can be easily done during system preparation via molecule IDs, only for reactive simulations this requires extra processing.
Nevertheless, there is no requirement that only one atom type must be used per element with ReaxFF. So it is still possible to group atoms by compound and you can also assign molecule IDs (they are just a label and have no physical effect on the simulation) in order to track groups of atoms.
Yes, I tried this, and fix reaxff/species command gave the results exactly I want.
One more question is about the molecule IDs, are those molecules IDs of each atom kept the same during the whole simulation? I mean, will new molecules IDs be generated if new molecules are generated? Or this mol IDs are just marks to track those atoms inside a molecule.
If a α molecule with molecule ID 1, and includes one C1, one O1, one H1 with molecule ID 1, after this α reacting with others and decomposing, C1 and O1 and H1 form other molecules, will those C1 O1 H1 still have molecule ID 1?