Dear Dr. Veld,
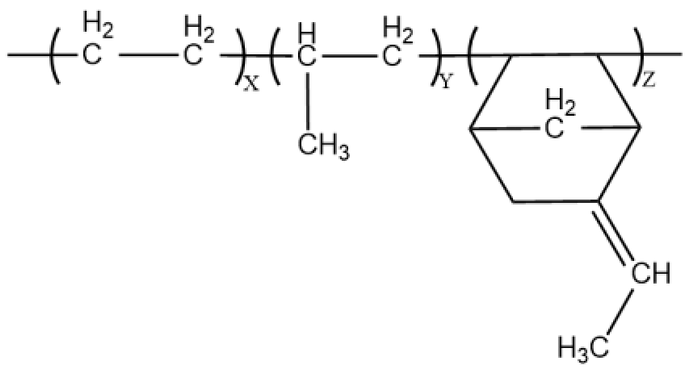
Hi! I am trying to creating a EPDM (Ethylene Propylene Diene Monomer) copolymer chain using EMC.
At the present, I am assuming there are only 1 ethyl, 1 propyl and 1 enb (5-Ethylidene-2-norbornene) in such a chain. Terminators are 2 methyls. My input script goes like this:
ITEM OPTIONS
field pcff
ntotal 44
mass true
replace true
density 0.87
ITEM END
ITEM GROUPS
ethyl *CC*,1,ethyl:2,2,propyl:1
propyl *CC(C)*,1,propyl:2,2,enb:1
enb CC=C1CC2CC1C(*)C2*,1,enb:2,2,ethyl:1 # the Diene Monomer part
methyl *C,1,ethyl:1,1,ethyl:2,1,propyl:1,1,propyl:2,1,enb:1,1,enb:2
ITEM END
ITEM CLUSTERS
poly random 1
ITEM END
ITEM POLYMERS
poly
100 ethyl,1,propyl,1,enb,1,methyl,2
ITEM END
This would work, but when visualizing the lammps data file (or pdb file) generated from this script, it seems like there are geometrical mistakes. So I am not sure if I put the asterisks at the right location in the SMILES string of enb monomer.
So I would like to know:
- If my ‘enb’ part in the ITEM GROUPS section comes with correct expression
- Is there any method that can assign ratios (x:y:z) for these three types of monomers? Because normally ethyl comes with largest portion, then propyl, and enb is the least.
- Can I create crosslinks between chains in EMC?
Thank you very much Dr. Veld.
Your advice and reply are highly appreciated! Thanks!