Dear relliott,
Thank you for your reply. I have figure out the install problem by using kim-api-collections-management install system EDIP… The reason is that I have no authority to access the usr/bin in the cluster.
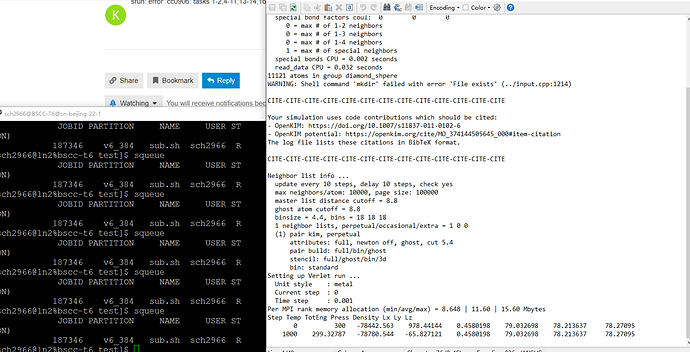
During my simulation, I find other problems. My lammps script, lammps log file, and cluster output information are as follows. According to the lammps log file, the kim potential involves 19 mutable parameters without abl potential parametes described in EDIP/C mode (zbl cutoff skin shift). How to use the EDIP/C mode? Another problem is the simulation terminates without run even one step. I tried in.kim-ex.melt from example/kim of lammps, and it runs ok.
Lammps script used:
variable file_name string start.data
variable tstep equal 0.001 # 0.5 fs timestep
variable start_temp equal 300
variable anneal_temp equal 1500
variable time_stage1 equal 10
variable time_stage2 equal 200
variable time_stage3 equal 1000 # anneal at this stage
variable time_stage4 equal 200 # cool the model down to start temp at this stage
variable dump_gap equal round(20/v_tstep) # dump one lammpstrj every dump_gap
variable fix_data_gap equal round(0.05/v_tstep) # output data every fix_data_gap
variable ptime equal stepv_tstep
variable Tdamp equal 100v_tstep
variable Pdamp equal 1000*v_tstep
variable runs_stage1 equal round(v_time_stage1/v_tstep)
variable runs_stage2 equal round(v_time_stage2/v_tstep)
variable runs_stage3 equal round(v_time_stage3/v_tstep)
variable runs_stage4 equal round(v_time_stage4/v_tstep)
#######################common settings############################
kim init EDIP_LAMMPS_Marks_2000_C__MO_374144505645_000 metal
boundary p p p
atom_style full
atom_modify map hash
read_data ${file_name}
mass * 12.0
kim interactions C
group diamond_shpere type 1
neighbor 2.0 bin
neigh_modify every 10 one 10000
###################define initial vel##############################
velocity all create ${start_temp} 4928459 mom yes rot yes dist gaussian
###################settings for thermo_style and dump#############
variable etotal equal etotal
compute 1 all displace/atom
compute 2 all stress/atom NULL
thermo_style custom step temp etotal press density lx ly lz
thermo 1000
shell mkdir results_info
shell cd results_info
fix info_print all print {fix_data_gap} "{ptime} ${etotal}" file energy.txt screen no
dump 1 all custom ${dump_gap} *.lammpstrj id type mol x y z c_1[4] c_2[4]
################################Constrain mmomentum of graphene###################
fix control2 diamond_shpere momentum 100 linear 1 1 1 angular
###################################Dynamic run#####################################
fix 1 all nvt temp {start_temp} {start_temp} {Tdamp} # x 0 0 {Pdamp} y 0 0 {Pdamp} z 0 0 {Pdamp}
#timestep {tstep}
run {runs_stage1}
The lammps log file
#######################common settings############################
kim init EDIP_LAMMPS_Marks_2000_C__MO_374144505645_000 metal
#=== BEGIN kim init ==========================================
units metal
neighbor 2.0 bin # Angstroms
timestep 1.0e-3 # picoseconds
This model has 19 mutable parameters.
No. | Parameter name | data type | extent
1 | eps | “Double” | 1
2 | BB | “Double” | 1
3 | beta | “Double” | 1
4 | sigma | “Double” | 1
5 | a | “Double” | 1
6 | aprime | “Double” | 1
7 | Z0 | “Double” | 1
8 | lambda0 | “Double” | 1
9 | lambdaprime | “Double” | 1
10 | gamma | “Double” | 1
11 | q | “Double” | 1
12 | flow | “Double” | 1
13 | fhigh | “Double” | 1
14 | alpha | “Double” | 1
15 | plow | “Double” | 1
16 | phigh | “Double” | 1
17 | Zdih | “Double” | 1
18 | Zrep | “Double” | 1
19 | c0 | “Double” | 1
#=== END kim init ============================================
boundary p p p
atom_style full
atom_modify map hash
read_data ${file_name}
read_data start.data
Reading data file …
orthogonal box = (-39.940409 -39.035132 -39.142455) to (39.092289 39.178505 39.128494)
6 by 4 by 4 MPI processor grid
reading atoms …
11121 atoms
Finding 1-2 1-3 1-4 neighbors …
special bond factors lj: 0 0 0
special bond factors coul: 0 0 0
0 = max # of 1-2 neighbors
0 = max # of 1-3 neighbors
0 = max # of 1-4 neighbors
1 = max # of special neighbors
special bonds CPU = 0.007 seconds
read_data CPU = 0.031 seconds
mass * 12.0
kim interactions C
#=== BEGIN kim interactions ==================================
pair_style kim EDIP_LAMMPS_Marks_2000_C__MO_374144505645_000
pair_coeff * * C
#=== END kim interactions ====================================
group diamond_shpere type 1
11121 atoms in group diamond_shpere
####################define the potential###########################
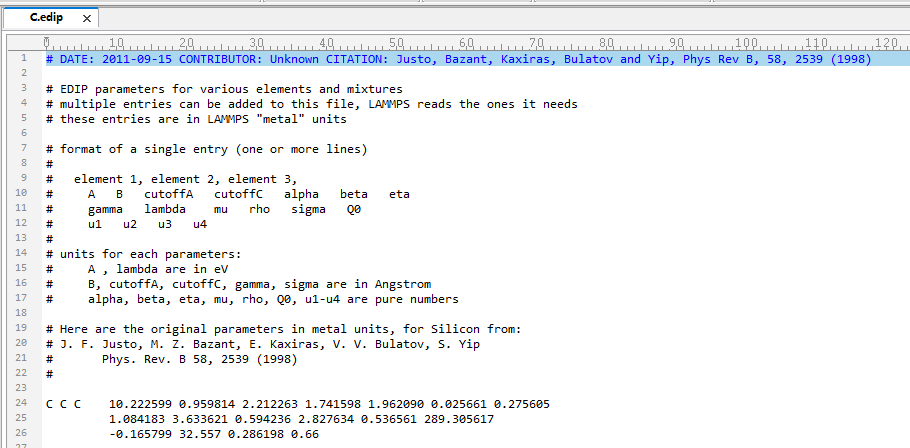
#pair_style edip
#pair_coeff * * C.edip C
#pair_style airebo 3.0 1 1 #
#pair_coeff * * CH.airebo C
neighbor 2.0 bin
neigh_modify every 10 one 10000
#neigh_modify exclude group graphene graphene check no
######################Energy minimization##########################
#min_style cg
#minimize 1e-20 1e-20 1000 100000
#write_data minimized.data
###################define initial vel##############################
velocity all create ${start_temp} 4928459 mom yes rot yes dist gaussian
velocity all create 300 4928459 mom yes rot yes dist gaussian
###################settings for thermo_style and dump#############
variable etotal equal etotal
compute 1 all displace/atom
compute 2 all stress/atom NULL
thermo_style custom step temp etotal press density lx ly lz
thermo 1000
shell mkdir results_info
WARNING: Shell command ‘mkdir’ failed with error ‘File exists’ (…/input.cpp:1214)
shell cd results_info
fix info_print all print {fix_data_gap} "{ptime} {etotal}" file energy.txt screen no
fix info_print all print 50 "{ptime} ${etotal}" file energy.txt screen no
dump 1 all custom ${dump_gap} *.lammpstrj id type mol x y z c_1[4] c_2[4]
dump 1 all custom 20000 *.lammpstrj id type mol x y z c_1[4] c_2[4]
################################Constrain mmomentum of graphene###################
fix control2 diamond_shpere momentum 100 linear 1 1 1 angular
###################################Dynamic run#####################################
fix 1 all nvt temp {start_temp} {start_temp} {Tdamp} # x 0 0 {Pdamp} y 0 0 {Pdamp} z 0 0 {Pdamp}
fix 1 all nvt temp 300 {start_temp} {Tdamp}
fix 1 all nvt temp 300 300 ${Tdamp}
fix 1 all nvt temp 300 300 0.1
#timestep {tstep}
run {runs_stage1}
run 10000
CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE
Your simulation uses code contributions which should be cited:
@Article{tadmor:elliott:2011,
author = {E. B. Tadmor and R. S. Elliott and J. P. Sethna and R. E. Miller and C. A. Becker},
title = {The potential of atomistic simulations and the {K}nowledgebase of {I}nteratomic {M}odels},
journal = {{JOM}},
year = 2011,
volume = 63,
number = 17,
pages = {17},
doi = {10.1007/s11837-011-0102-6}
Cluster output file:
LAMMPS (30 Jul 2021)
OMP_NUM_THREADS environment is not set. Defaulting to 1 thread. (…/comm.cpp:98)
using 1 OpenMP thread(s) per MPI task
Reading data file …
orthogonal box = (-39.940409 -39.035132 -39.142455) to (39.092289 39.178505 39.128494)
6 by 4 by 4 MPI processor grid
reading atoms …
11121 atoms
Finding 1-2 1-3 1-4 neighbors …
special bond factors lj: 0 0 0
special bond factors coul: 0 0 0
0 = max # of 1-2 neighbors
0 = max # of 1-3 neighbors
0 = max # of 1-4 neighbors
1 = max # of special neighbors
special bonds CPU = 0.007 seconds
read_data CPU = 0.031 seconds
11121 atoms in group diamond_shpere
WARNING: Shell command ‘mkdir’ failed with error ‘File exists’ (…/input.cpp:1214)
CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE
Your simulation uses code contributions which should be cited:
CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE-CITE
Neighbor list info …
update every 10 steps, delay 10 steps, check yes
max neighbors/atom: 10000, page size: 100000
master list distance cutoff = 8.8
ghost atom cutoff = 8.8
binsize = 4.4, bins = 18 18 18
1 neighbor lists, perpetual/occasional/extra = 1 0 0
(1) pair kim, perpetual
attributes: full, newton off, ghost, cut 5.4
pair build: full/bin/ghost
stencil: full/ghost/bin/3d
bin: standard
Setting up Verlet run …
Unit style : metal
Current step : 0
Time step : 0.001
srun: error: cc0906: tasks 0,3,12,15,80,83,92,95: Segmentation fault
srun: First task exited 60s ago
srun: StepId=187275.0 tasks 1-2,4-11,13-14,16-79,81-82,84-91,93-94: running
srun: StepId=187275.0 tasks 0,3,12,15,80,83,92,95: exited abnormally
srun: launch/slurm: _step_signal: Terminating StepId=187275.0
srun: Job step aborted: Waiting up to 62 seconds for job step to finish.
slurmstepd: error: *** STEP 187275.0 ON cc0906 CANCELLED AT 2022-03-22T15:23:13 ***
srun: error: cc0906: tasks 1-2,4-11,13-14,16-79,81-82,84-91,93-94: Killed