Since the feature in LAMMPS is new, I am willing to refactor or rewrite it.
While I started with trying to directly translate LAMMPS regions into VMD graphics primitives, I am slowly moving toward doing a full triangulation in LAMMPS and then output just triangles or triangles with norm vectors at the tips for smoothly rounded surfaces. This way, I can visualize the cone region and the cylinder becomes just a special case of a cone, same for ellipsoid and sphere. The block and prism styles are made from pairs of triangles for each side and plane is still in development, but could be built from 4 triangles around the center point.
Intersections and unions are - at the moment - too hard a problem. They are easy to handle in LAMMPS since for the use of regions, LAMMPS looks at atoms, i.e. whether the coordinates fit into all or one of the combined regions. Visualizing them is harder, but people can still visualize the constituent regions and by switching colors one could tell them apart.
The more I think about it, the more I am leaning toward creating a generic class that does the triangulation for all supported regions and stores the information internally. Then there could be multiple commands to output this information in different file formats. So not just VMD script, but also, say, STL. Then there would be multiple commands, region2XXX that will output the triangulation in a different formats. That would also elegantly solve the issue of moving or rotating regions, since that transformation can be performed separate (and only if needed) from generating the triangulation. The latter could also include flags, whether a region has a variable shape or is fixed and then the triangulation only needs to be repeated for those with a variable shape.
That would be problematic for anything that uses variables, i.e. shape, movement, or rotation.
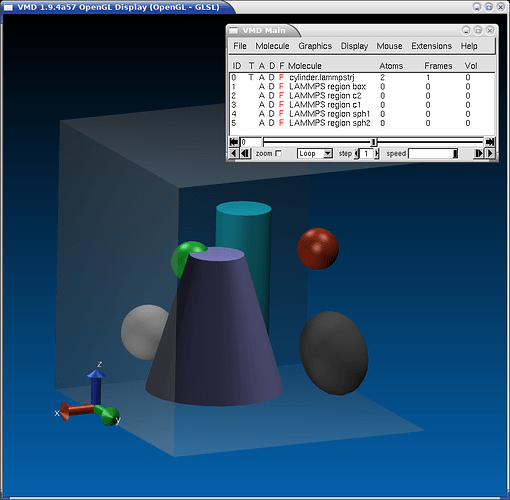
My current feeling is that having different back-ends would be more suitable, since it allows to “optimize” how the data is organized for the visualization tool/strategy at hand. Since most regions are static, I have started with a simple command that creates a single file for a single timestep. In VMD I would have to use VMD/Tcl scripting and for visualizing a trajectory, I would need write a script that stashes away the triangulation data into an “array of structs” and then connect the coordinate dump file step with the matching triangulation data. The cleanest way in VMD to handle this (and allow people to individually turn region visualizations on or off) would be to create an empty “molecule” for each region and then add the graphics primitives to that. This is what I currently have. and then the VMD main window would look like this.
By clicking on the “D” character in each line for a “molecule” the region can be hidden or displayed and the “molecule” name includes the region ID.
And below is the VMD input to generate this. I then run ./lmp -in in.input and then visualize with vmd -e cylinder.vmd. The “command” keyword in the region2vmd command, allows me to insert custom VMD script commands, so that the resulting .vmd file can be used as a VMD state file and loaded accordingly and will load the dumped coordinates as well. Of course, a region2ovito command could handle this in a way that is optimal for OVITO.
units real
region box block 0 20 0 20 0 20 open 1 open 4
create_box 2 box
mass * 1.0
region sph1 sphere 15 5 5 2
region sph2 ellipsoid 5 15 5 2 4 3
create_atoms 1 single 5 12 12
create_atoms 2 single 15 12 12
variable xxx equal -2.0
region c1 cylinder z 10 10 2 5 15
region c2 cone z 14 14 5 2 5 15 move NULL NULL v_xxx
run 0 post no
write_dump all atom cylinder.lammpstrj
region2vmd cylinder.vmd command "mol default style VDW" &
command "set oldtop [mol new cylinder.lammpstrj waitfor all autobonds off]" &
command "axes location LowerLeft" region box &
material Diffuse color iceblue region c2 color cyan region c1 &
color white region sph1 color gray region sph2
I want to start by writing a command for a single static visualization to be used in combination with a file generated by write_dump or write_data, and then develop a corresponding dump style on top of that. I see that there could be at least four back ends: VMD, OVITO, STL, and VTK.
Since we already have an option for exporting VTK, the logical step would be to expand that dump style. For STL output, we would have to enforce the one file per timestep option with embedding a star character in the filename. VMD will have to use a mechanism similar to what is used in my 20+ year old tutorial on dynamic data with VMD: Visualization and Analysis of Quantum Chemical and Molecular Dynamics Data with VMD. In this case a region2vmd dump style would have to be given the dump ID of a trajectory dump with coordinate data, so the VMD script can be synchronized with it. It is very likely that OVITO needs the data in a different style (unless you want to write a parser for VMD script commands and translate them to OVITO), so it would be up to you define it. If you prefer a custom format, it would be straightforward to follow other LAMMPS dump style conventions with “ITEM: XXX” tags and the corresponding information following.
My plan is to start the refactoring according to what I have outlined here soon, I can make the branch public and then we can work on the details and the various back ends together, either by you commenting on the code and making suggestions for (small) changes or by submitting pull requests for larger changes.
Please let me know what you think about my plans.