Hi,
I’m using ovito-pro-3.3.4 and have tested the grain segmentation algorithm on a system containing grains of Cubic Diamond and Hexagonal Diamond structures, and it seems “handle_stacking_faults=True” isn’t working for me.
Does this algorithm handle diamond stacking faults, or is it for HCP/FCC stacking faults only?
The structure file I provided (correction: apologies but the forum won’t let me upload files as I am a new user) is a test system with nearly stacking disordered cubic and hexagonal diamond. So should return one Grain for the entire system. However it returns 7 grains, due to cubic and hexagonal regions being identified as their own grains.
Code:
topoFILE=’./test.pdb’
create pipeline
pipeline = import_file(os.path.abspath(topoFILE))
PTM = PolyhedralTemplateMatchingModifier(color_by_type=False, output_orientation=True)
PTM.structures[PolyhedralTemplateMatchingModifier.Type.CUBIC_DIAMOND].enabled = True
PTM.structures[PolyhedralTemplateMatchingModifier.Type.HEX_DIAMOND].enabled = True
pipeline.modifiers.append(PTM)
ALG = GrainSegmentationModifier.Algorithm.GraphClusteringAuto
grains = GrainSegmentationModifier(algorithm=ALG,color_particles=True,handle_stacking_faults=True, min_grain_size=100,orphan_adoption=False)
pipeline.modifiers.append(grains)
compute
data = pipeline.compute()
n_grains = len(np.unique(data.particles[‘Grain’]))
print(‘Number of grains: {}’.format(n_grains))
Note:
I’ve also tried GrainSegmentationModifier.Algorithm.GraphClusteringManual with varied merge_thresholds. Above ~ 20 it returns 6 grains. Testing all the way up to 100’000s shows no variation beyond this though.
I’d be very grateful for any help on this!
Many thanks,
Michael
Hi Michael,
I would like to take a look at your input structure myself and will consult Peter Mahler Larsen if necessary. In principle, stacking faults in diamond structures should be handled by the algorithm. Peter extended the stacking fault handling to this crystal structure case in one of the recent development cycles, I thought. But I need to verify this myself.
I’ve raised your trust level in the forum. You should now be able to attach files to your posting, hopefully – I am rather new as a moderator of this forum myself. Please try again uploading your structure file. Thanks.
-Alex
Hi Alex,
Thanks for the prompt response. The PDB file is 25 Mb in size, which is too big to upload here apparently so I have put it on a dropbox link here: https://www.dropbox.com/s/ntnrhnpv7sllltl/test.pdb?dl=0
If there’s any issues with this just let me know.
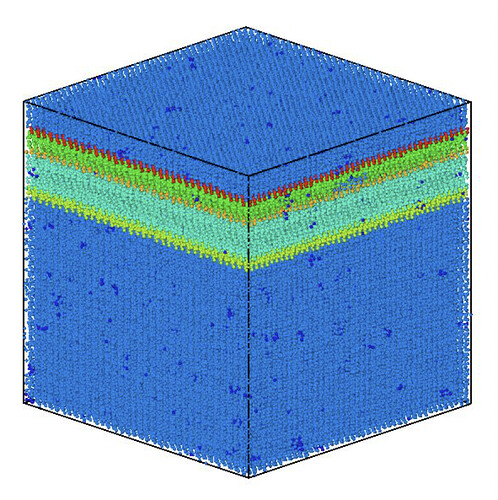
I’ve also attached two images of the system, one coloured by polytype (hex or cubic) and one coloured by grain, to help show the issue.
Thanks for providing the simulation file. I am going to investigate this behavior of the algorithm together with Peter Mahler Larsen, who implemented the handling of stacking faults. It may be that in this case the algorithm is indetermined whether this input represents a hexagonal diamond crystal with cubic stacking faults or rather a cubic crystal with hexagonal stack fault regions embedded.
Peter has fixed this issue in the grain segmentation algorithm. Due to a recent regression the handling of stacking faults of arbitrary thickness didn’t work as expected.
I’ve released a new update of OVITO containing the fix. Please download and install OVITO 3.3.5 and try it again. We appreciate your feedback regarding the experimental grain segmentation algorithm.
Thank you both for addressing this so quickly! I’ve tried the new version and it works now.