Hello, dear pymatgen users! Recently, I’ve encountered some difficulties while converting CIF files containing fractional occupancy into POSCAR files for VASP calculations.
When faced with a CIF file with fractional occupancy, I used the following simple script for conversion (the script is attached at the end, pymatgen_cif2poscar_1.0.py):
from pymatgen.io.vasp import Poscar
from pymatgen.core import Structure
file_name="Ca0.134Zr0.866O1.7-icsd-60604_save_diff_redu.cif"
structure = Structure.from_file(file_name) # Attention path
#----------------------------------------
# The ICSD file, i.e. cif, is considered here
# Convert cif file to poscar file
#1.Only the first three coordinates in the cif file are considered to avoid possible accuracy problems
rounded_sites = {}
for site in structure:
# Round the coordinates to three decimal places
rounded_coords = tuple(round(x, 3) for x in site.frac_coords)
if rounded_coords not in rounded_sites:
rounded_sites[rounded_coords] = {}
for element, occupancy in site.species.items():
if element in rounded_sites[rounded_coords]:
rounded_sites[rounded_coords][element] += occupancy
else:
rounded_sites[rounded_coords][element] = occupancy
#2. Create a simplified structure
simplified_sites = []
for coords, species in rounded_sites.items():
# Chose the most important element
most_likely_species = max(species.items(), key=lambda item: item[1])[0]
simplified_sites.append((most_likely_species, coords))
#3. Create a new Structure object
lattice = structure.lattice
elements = [site[0] for site in simplified_sites]
coords = [site[1] for site in simplified_sites]
simplified_structure = Structure(lattice, elements, coords)
#----------------------------------------
#The simplified Structure object is converted to poscar file and output
poscar = Poscar(simplified_structure)
poscar.write_file(f"0-{file_name}.vasp")
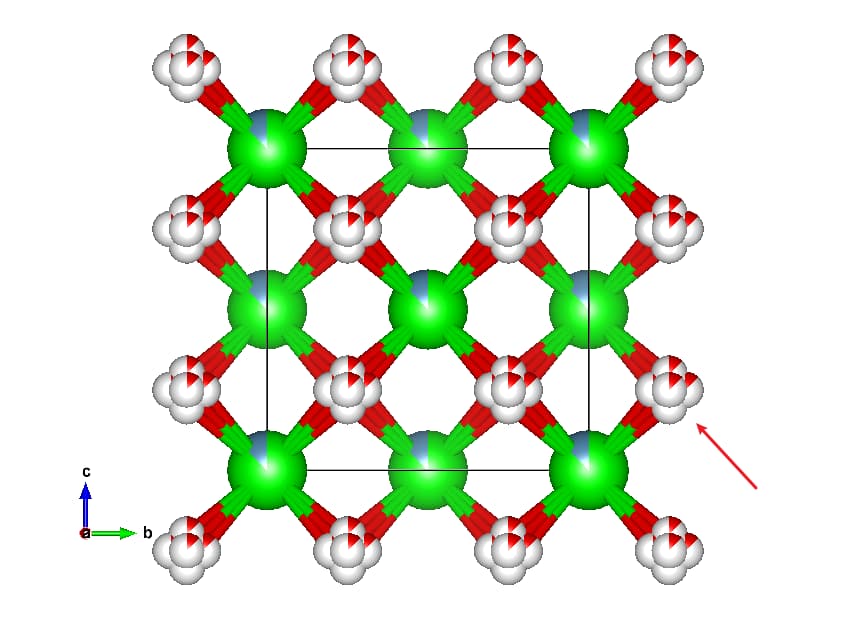
In this script, I simply select the element with the highest occupancy at a fractional occupancy site. For example, in CeZr.cif (this file is also attached at the end):
The script converts the Ce/Zr fractional occupancy site to Ce only.
However, this approach leads to some issues:
Issue 1:
Since the generated POSCAR only considers the element with the highest occupancy, this leads to a significant deviation in the stoichiometry of the POSCAR from the actual CIF values. In the example above, this approach might even exclude elements!
Issue 2:
Even if the site contains only one possible element, like oxygen in CaZrO.cif, numerous uncertain positions not only make the POSCAR’s stoichiometry inaccurate but also result in an unrealistic structure.
For Issue 1, I’m considering using a supercell approach to solve this. I found EnumerateStructureTransformation on the pymatgen API Documentation. As for Issue 2, I don’t have any ideas.
Are there any good solutions for the above two issues? I’d appreciate more detailed guidance. Thank you very much!
pymatgen_cif2poscar_1.0.py (2.4 KB)
CeZr.cif (2.0 KB)
CaZrO.cif (5.8 KB)