Hi, I am practice the code was given in the article " A high throughput framework for determining adsorption energies on solid surface" by Montoya.
I have an error and I dont understand the error msg. Could anyone help me with this?
Thank you so much!
Hi Ngan, I’ll need some time to dig into the specific issue with this code. In the meantime, can I confirm that you’re using the latest version, which is maintained here?
https://matgenb.materialsvirtuallab.org/2018/07/24/Adsorption-on-solid-surfaces.html
Hi Joseph,
Thank you for the response. I fixed the script and it worked. I used the supplemental document which came with the paper, I guess i need to specify “all” in order for it to work.
I have another question for this topic. I hope you can answer when you have some time.
This is the code I would like to run where I want to generate different miller indexes for the same metal. When I ran the code, somehow it only print the last structure of each metal instead of plotting all.
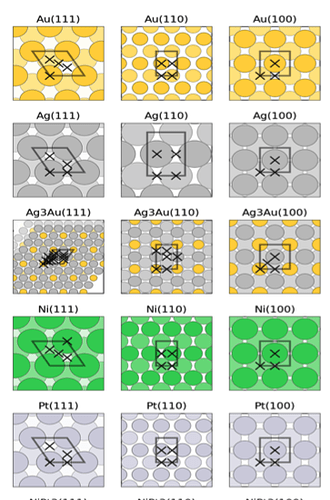
I would like to plot it out like this example so I can compare. Do you know how to fix this issue?
In addition, I use Poscar in vasp.input to write all the structures to POSCAR file using the script below
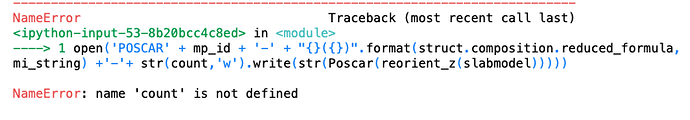
open(‘POSCAR’ + mp_id + ‘-’ + “{}({})”.format(struct.composition.reduced_formula, mi_string) +’-’+ str(count,‘w’).write(str(Poscar(reorient_z(slabmodel)))))
And I got this message
Do you have other suggestion to write all the structure to POSCAR file using few lines of code? Im new to Python and Pymatgen so I dont know how to fix this error
Thank you again for your time!
Ngan Huynh
For the first question, you’re using a python dictionary which can only have one value per unique key, so only the last one is kept in the data structure resulting from the code. You should be able to use a list of tuples if you want multiple pairs with the same material, something like:
slab_data = [
("mp-134", (1, 0, 0)),
("mp-134", (1, 1, 1)),
...
]
# Note no .items() here
for n, (mp_id, m_index) in enumerate(mats):
struct = mpr.get_structure_by_material_id(mp_id)
...
For the second, you should be able to use the Structure.to method attached to Slab, since it’s subclassing structure, with an appropriate iterator for the slab objects that are accumulated in your above script.
for slab in slabs:
slab.to(filename="POSCAR_{}-{}".format(
slab.composition.reduced_formula, slab.miller_index))
Thank you so much for your time and your help!
It saves me a lot of time since I usually do the POSCAR manual and the adsorbate_sitefinder for some of the oxides i study have 10+ structure
Appreciate your time
Ngan Huynh
Hi Joseph,
I have another question relate to this topic.
After I added the adsorbate to the structure and use the code:
“for slab in slabs: slab.to(filename=“POSCAR_{}-{}”.format( slab.composition.reduced_formula, slab.miller_index))”
It only print one structure instead of 3 or 4 structures as shown in the find_adsorption_sites figure.
Here is my code
I think the for-loop at the end of the script has some error but I am not sure how to place the index of each position to write the POSCAR file.
Thank you for your time!
Kelly
Hi Kelly, can you post the text of your code? If you want it formatted as a code block, you can enclose it in triple backticks ```, e. g.
print("Hello world!")
This makes it easier to copy/paste the contents. Also, if you’re encountering an error, please paste the text of the error.