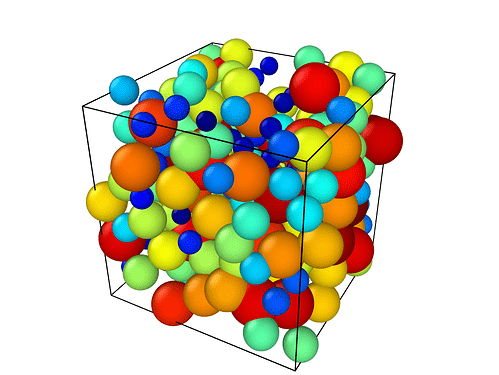
Hi, Currently I am working on creating a 3D system of X number of spheres with random radius between 2 different values. I have consulted everything about this, but I can’t solve this myself and hope someone can help me with this. I will briefly explain what I have done so far. I’ve defined an input script with all sections: I use lennard jones potential and atom_style sphere, because I’m interested in the positions and diameter.
Then I defined properties as follows:
variable rmin equal 1.0 # minimum value for the radius
variable rmax equal 3.0 # maximum value for the radius
variable rdifference equal {rmax}-{rmin}
variable radius equal {{rmin}+{rdifference}*random()}
create_box 1 simulation_box
create_atoms 1 random ${N} 12345 NULL overlap 1.0 maxtry 1
group all type 1
set group all diameter ${radius}
When running the simulation, the random generator function “random()” does not work and I get “ERROR: Variable radius: Invalid syntax in variable formula”. I don’t understand why? So then I tried to solve this with the following line: variable radius equal ceil(random(0,3,1)), where 0 is minimum value, 3 is maximum value, and 1 represents seed (which I don’t understand by the way?).
This works, but all the spheres keep the same constant radius. In addition, the value remains the same even though I run it several times.
Then I tried a loop:
Loop over the spheres and create them
Label loop_spheres
variable i loop {N} # number of iterations
variable radius equal ceil(random(1,3,1))
variable x equal ceil(random(1,20,1))
variable y equal cell(random(1,20,1))
variable z equal cell(random(1,20,1))
create_atoms 1 sphere {x} {y} {z} ${radius}
next i
jump SELF loop_spheres
The “create_atoms …” line gives the following error: Unknown create_atoms command option sphere. If I delete this line nothing happens, so the radius and positions do not change!
Finally, I created a data_file.txt myself with python script and used the following format for atom_style sphere:
atom-ID atom-type diameter density x y z
1 1 5.48 1 1 9 1
2 1 4.40 1 7 7 0
3 1 2.10 1 11 19 1
4 1 4.03 1 17 4 3
5 1 2.74 1 5 2 9
6 1 5.32 1 6 17 13
7 1 2.49 1 16 12 13
8 1 2.21 1 19 10 10
9 1 3.54 1 2 5 11
10 1 2.06 1 4 13 9
After the initialization, defining of sphere properties, system definition and simulation settings, I added the following line command to read the file: read_data data_read_in.txt add append
with data_read_in.txt name of file and ‘add append’ to indicate that simulation box is defined.
Error that now follows:
ERROR: Unexpected end of data file (src/read_data.cpp:1968)
Last command: read_data data_read_in.txt add append
I don’t know what else to do and hope someone can help me with this? I would also like to know when the file is read. How do I then link those values to the variable radius and x,y,z positions to create randomly sized spheres?