Dear user,
I looked into your problem a little more in depth. The problem is caused by how VMD creates a carbon nanotube. The resulting structure consists of as many residues as there are atoms, which causes problems for EMC when recognizing groups and corresponding bond valences. An added complication forms the ends of the nanotube, which are not terminated by hydrogens. This can be circumvented by creating a unit cell for a carbon nanotube and use this as a template for creating a continuous tube embedded in a polymer melt. The unit cell is given by
unit.pdb (3.3 KB)
unit.psf (4.2 KB)
These two files need to be imported in order to embed them in a polymer. Here the polymer is chosen to be polyethylene. An EMC Setup script would look like
ITEM OPTIONS
replace true
field basf
#number true
ntotal 10000
moves_cluster active=true
direction x
density 1
emc_execute true
ITEM END
ITEM GROUPS
ethyl *CC*, 1,ethyl:2
methyl *C, 1,ethyl:1, 1,ethyl:2
ITEM END
ITEM CLUSTERS
molecule import, name="unit", type=tube, mode=pdb, percolate=true, &
tighten=10, ncells=2:2:10, exclude=contour, direction=z
polymer alternate,1
ITEM END
ITEM POLYMERS
polymer
1 ethyl,10,methyl,2
ITEM END
The only problem is, that the current version online does not support type=tube. Therefore, I included the resulting build.emc,
build.emc (6.2 KB)
which should be working with the new online EMC version (v9.4.4 of 20230801, just a newer updated version, to be downloaded from here). The attached build.emc builds four continuous nanotubes and embeds them in a PE melt. Changes can be made by
emc.sh -ntotal=n -nx=mx -ny=my -nz=mz -ltighten=l build.emc 2>&1 | tee build.out
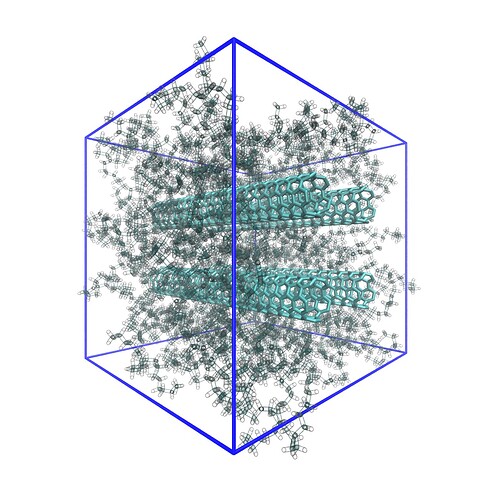
Values were chosen to be n=10000, mx=2, my=2, mz=10, and l=10, as reflected by the above .esh example. Note that ltighten controls the spacing between the nanotubes. The following represents the resulting structure: