Hello all,
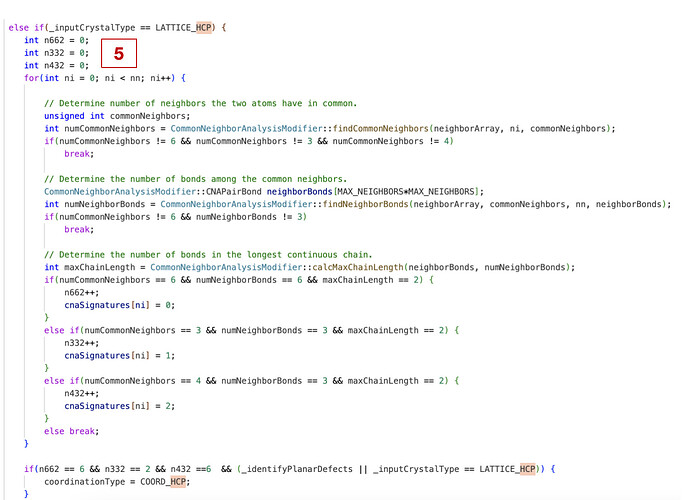
When using ovito to recognize dislocations in crystals, the dislocations are not recognized due to the variable c/a value of the HCP structure. For example, I want to recognize the dislocations in WC, but the c/a value of WC is much different from the standard. I tried to partially modify the source code of the DXA part in OVITO (as shown in the figure), and there is a difference in the recognition result before and after the modification but the difference is not big. I would like to ask for advice on how I should solve the problem of not recognizing all dislocations in WC.
When using DXA, there is a first step where Common Neighbor Analysis (CNA) is used to determine the crystal structure of each atom.
I assume that CNA does not work for your WC structure. You can easily test this by adding just the CNA modifier into your pipeline and checking the fraction of HCP atoms.
If this is indeed correct, you would have to either modify CNA to correctly identify these kinds of distorted HCP structures. Alternatively, if you are working with a single crystal, you can use the Affine Transformation modifier to first strain your system to a c/a ratio of 1.6 before applying DXA.
Thank you so much. I have another question, using Polyhedral template matching can recognize the HCP structure of WC, can I use PTM to work with DXA. I tried to use PTM to recognize the structure first and then use DXA to recognize dislocation , but DXA still can’t recognize the structure and dislocations.
DXA performs its own structure identification internally, based on CNA but with additional features. It does not depend on any structure identification algorithm from the upstream pipeline. Therefore, to use a different structure identification algorithm, you would need to update the DXA implementation.
Thanks a lot !
In order to recognize the dislocations in WC with non-standard HCP structures, I have attempted to modify the DXA part of the source code, and have now learned that I need to modify the StructureAnalysis.cpp file first, so that DXA can recognize the structure of the WC. I have already modified the vectors of the nearest-neighbor atoms and next-nearest-neighbor atoms, and now I have a question, I found two cutoff radius in this file, as shown in the red box marked in the figure, what are the similarities and differences between these two values, and what modifications do I need to make so that they can be applied to recognize the WC structure.
These cutoffs should correspond to equations 4 and 6 in Stukowski, A., Structure identification methods for atomistic simulations of crystalline materials. Modelling and Simulation in Materials Science and Engineering vol. 20 045021 (2012).
Thank you for your continued patience and guidance throughout this process. I’m writing to consult regarding some adjustments I’ve made to the StructureAnalysis.cpp code in an effort to identify WC structures with non-standard c/a ratios through dislocation extraction algorithm (DXA). Here’s an overview of the key modifications implemented:
- Implemented atomic coordinate definitions for WC structure including:
- 6 nearest neighbors
- 2 second-nearest neighbors
- 6 third-nearest neighbors
- Adjusted cutoff radius to the mean value between third and fourth nearest neighbor distances
- Configured analyzed atom count parameter to 14
- Modified localcutoff parameters adopting BCC structure’s weighted approach
- Updated CNA criteria to: (n662 == 6 && n332 == 2 && n432 == 6)
However, post-compilation testing indicates the DXA module still fails to recognize WC structures. Would you be available to:
- Review whether existing modifications require optimization
- Suggest any additional parameters needing adjustment
- Identify potential conflicts in current implementation?
I’d be grateful for your professional insights when convenient. Please let me know if you require any implementation details or test cases for further analysis.
Hi, thanks for sharing this update.
In principle this seems to be going into the right direction - as far as I can tell from here. But I have not double checked the calculation of the adaptive cutoff radius, which you have implemented. Also, I couldn’t verify the CNA indices 662, 332 and 432, which you determined, without having a reference structure for testing.
Could you please explain a bit more why you decided to use the 3 nearest neighbor shells in the CNA calculation? Is this choice a particularly good one for the WC structure?
If you would like us to help with the development and validation of your modifications, it would be necessary to give us access to the source code, not just screenshots. Did you fork the OVITO code repository and is your fork publicly accessible somewhere?
Thank you for your guidance. I’ve attached the revised StructureAnalysis.cpp and would appreciate your feedback on potential issues.Here’s a brief explanation of the questions you raised:
- WC Structure Parameters:
Non-standard c/a ratio = 0.973 (a=2.917Å, c=2.839Å)
Neighbor distances:
• 1st (W-C): 2.203Å
• 2nd: 2.839Å
• 3rd: 2.917Å
• 4th: 4.070Å
Cutoff radius set to 3.494Å (average of 3rd/4th neighbors)
Coordinate scaling:
a = sqrt(2.0)/2.0 → Actual lattice conversion:
2.917/(√2/2) = 3.494/cutoff , cutoff = 0.85 - CNA Index Validation
The attached Python script calculates CNA indices using ckDTree for neighbor detection. Could you advise if this neighbor-traversal logic properly determines CNA indices and recommended methods to verify indices against WC structure. - For the WC structure, the coordination consists of 6 first-nearest neighbors, 2 second-nearest neighbors, and 6 third-nearest neighbors. When using a smaller cutoff radius that only accounts for the first-nearest neighbors, the CNA indices I obtained showed 000 patterns. Due to the nearly identical distances between second-nearest (2.839Å) and third-nearest (2.917Å) neighbors, I’ve extended the analysis to include third-nearest neighbors in the CNA calculation to better capture the structural signature.
Attached are three key files:
- The WC structure file WC.cif
- Python code for calculating CNA indices cna_HCP.py
- My modified version of the source code StructureAnalysis.cpp
I would appreciate your guidance on whether this approach appropriately addresses the characteristic coordination environment of WC structures. I’d be happy to provide test cases or discuss further details.
WC.cif (828 Bytes)
cna_HCP.py (4.3 KB)
StructureAnalysis.cpp (56.6 KB)
Thanks for providing more details.
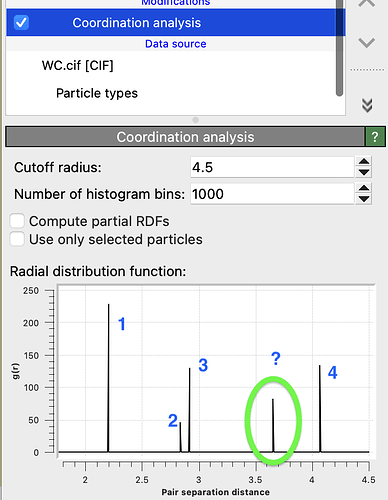
Is there a neighbor shell missing from this list?
I have used the Coordination Analysis modifier of OVITO to calculate the shell structure, which shows an additional shell at r=3.65481:
Furthermore, this calculation likely is wrong:
localCutoff = localScaling * (1.0f + sqrt(2.0f)) * 0.5f * 1.87;
localScaling is the nearest neighbor distance (2.203Å), but then the formula doesn’t yield the average of the 3rd & 4th neighbor distance.
Thank you very much for your patient explanation and guidance.
We have confirmed that the distance of the fourth nearest neighbor atom is indeed 3.65481, and identified an issue with the localcutoff parameter. After making the necessary adjustments to the code, we have successfully recognized the HCP structure in WC. Attached please find the modified files for your reference. May I kindly inquire whether the successful identification of the WC structure using DXA indicates that our code modifications are correct? Your professional insight would be greatly appreciated.
StructureAnalysis.cpp (56.6 KB)
Congratulations on getting this to work. Yes, the CNA step now seems to recognize the hcp structure successfully. It remains to be seen whether the subsequent DXA step is able to extract the dislocation defects found in such a crystal.
Do you have a simulation of WC with defects that you can use to test the dislocation identification?
In the meantime I have also explored other ways of extending the CNA to recognize the WC structure. I came up with a different approach, which is basically a preprocessing step. Instead of modifying the CNA algorithm itself and implementing new signatures for the 14 neighbors, my solution first identifies the c-axis direction of the crystal first to compute an estimate for the c/a ratio. Using this information, it applies a uniaxial counter-deformation to the atomic neighborhood to produce a local structure with an approximately ideal c/a ratio. This structure can then be fed into the standard CNA procedure to identify the hcp phase.
I have generated test builds of OVITO so you can try this alternative approach and compare it to your solution. The source code for my modified version can be found in this development branch.
ovito-basic-3.12.1-dev-8bc0298e-x86_64-dxa-hcp-extension.tar.xz
ovito-basic-3.12.1-dev-8bc0298e-macos-arm64-dxa-hcp-extension.dmg
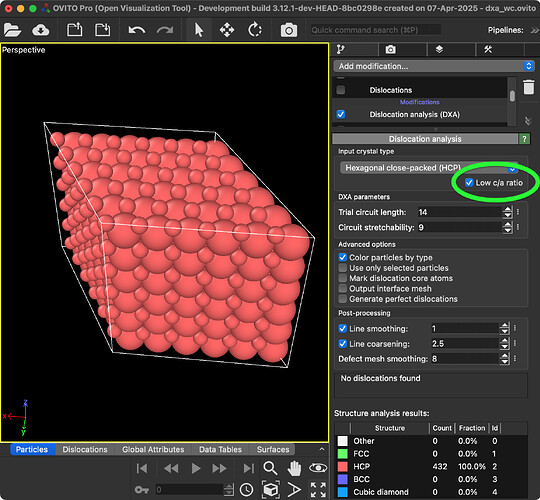
The “Low c/a ratio” option activates the described preprocessing step.
Thank you for your reply.
I tried to use your update to successfully identify the structure of the polycrystalline WC as well as the dislocations, and the difference between the code we modified and the results identified by your update is not too bad.