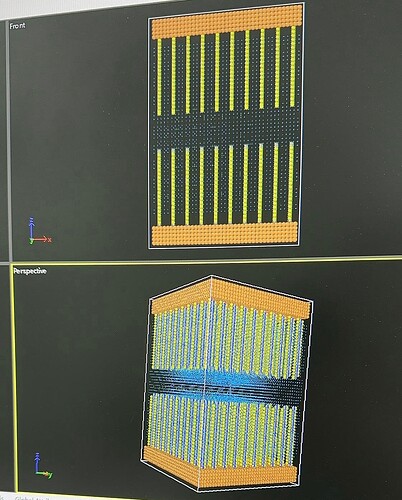
Dear all:
- My model is organic molecules grafted between the upper and lower plates, with NaCl solution in the middle! Organic molecules and solvent beads are partially charged!The system contains about a hundred thousand beads!
- If I don’t add the long range Coulomb interaction called coul/long, the whole system takes only ten minutes to complete the calculation!!If the long-range Coulomb interaction is added, the calculation speed becomes quite slow, which increases to about a month!!
3.My question is how to adjust my in file to reduce the computation time! - Here is my in file:
units lj
dimension 3
boundary p p p
neighbor 10 bin
neigh_modify delay 10 every 2 check yes
atom_style full
pair_style hybrid/overlay dpd 1.0 1.0 343587 coul/long 3.0
bond_style harmonic
comm_modify vel yes cutoff 3.5
kspace_style ewald 1.0e-4
read_data data.200000
group wall type 5
group head type 1
group C type 2
group N type 3
group S type 4
group W type 6
group NaCl type 7 8
group poly type 1 2 3 4
group ss union head wall
set group N charge 1
set group S charge -1
pair_coeff 1 1 dpd 25.0 4.5 1.0
pair_coeff 1 2 dpd 25.0 4.5 1.0
pair_coeff 1 3 dpd 25.72 4.5 1.0
pair_coeff 1 3 coul/long
pair_coeff 1 4 dpd 25.05 4.5 1.0
pair_coeff 1 4 coul/long
pair_coeff 1 5 dpd 800 4.5 1.0
pair_coeff 1 6 dpd 31.21 4.5 1.0
pair_coeff 1 7 dpd 31.21 4.5 1.0
pair_coeff 1 7 coul/long
pair_coeff 1 8 dpd 31.21 4.5 1.0
pair_coeff 1 8 coul/long
pair_coeff 2 2 dpd 25.0 4.5 1.0
pair_coeff 2 3 dpd 25.72 4.5 1.0
pair_coeff 2 3 coul/long
pair_coeff 2 4 dpd 25.05 4.5 1.0
pair_coeff 2 4 coul/long
pair_coeff 2 5 dpd 800 4.5 1.0
pair_coeff 2 6 dpd 31.21 4.5 1.0
pair_coeff 2 7 dpd 31.21 4.5 1.0
pair_coeff 2 7 coul/long
pair_coeff 2 8 dpd 31.21 4.5 1.0
pair_coeff 2 8 coul/long
pair_coeff 3 3 dpd 25 4.5 1.0
pair_coeff 3 3 coul/long
pair_coeff 3 4 dpd 25.78 4.5 1.0
pair_coeff 3 4 coul/long
pair_coeff 3 5 dpd 800 4.5 1.0
pair_coeff 3 5 coul/long
pair_coeff 3 6 dpd 31.35 4.5 1.0
pair_coeff 3 6 coul/long
pair_coeff 3 7 dpd 31.35 4.5 1.0
pair_coeff 3 7 coul/long
pair_coeff 3 8 dpd 31.35 4.5 1.0
pair_coeff 3 8 coul/long
pair_coeff 4 4 dpd 25.0 4.5 1.0
pair_coeff 4 4 coul/long
pair_coeff 4 5 dpd 800 4.5 1.0
pair_coeff 4 5 coul/long
pair_coeff 4 6 dpd 33.03 4.5 1.0
pair_coeff 4 6 coul/long
pair_coeff 4 7 dpd 33.03 4.5 1.0
pair_coeff 4 7 coul/long
pair_coeff 4 8 dpd 33.03 4.5 1.0
pair_coeff 4 8 coul/long
pair_coeff 5 5 dpd 25.0 4.5 1.0
pair_coeff 5 6 dpd 800 4.5 1.0
pair_coeff 5 7 dpd 800 4.5 1.0
pair_coeff 5 7 coul/long
pair_coeff 5 8 dpd 800 4.5 1.0
pair_coeff 5 8 coul/long
pair_coeff 6 6 dpd 25.0 4.5 1.0
pair_coeff 6 7 dpd 25.0 4.5 1.0
pair_coeff 6 7 coul/long
pair_coeff 6 8 dpd 25.0 4.5 1.0
pair_coeff 6 8 coul/long
pair_coeff 7 7 dpd 25.0 4.5 1.0
pair_coeff 7 7 coul/long
pair_coeff 7 8 dpd 25.0 4.5 1.0
pair_coeff 7 8 coul/long
pair_coeff 8 8 dpd 25.0 4.5 1.0
pair_coeff 8 8 coul/long
mass 1 1.0
mass 2 1.0
mass 3 1.0
mass 4 1.0
mass 5 1.0
mass 6 1.0
mass 7 1.0
mass 8 1.0
bond_coeff * 180 0.8
velocity all create 1.0 4928459 dist gaussian
fix 1 ss setforce 0.0 0.0 0.0
velocity ss set 0.0 0.0 0.0
timestep 0.001
thermo 10
thermo_style custom step dt time atoms temp press pe ke etotal evdwl ecoul epair elong enthalpy vol density
thermo_modify flush yes lost ignore lost/bond ignore
dump 1 all custom 100 dump.lammpstrj.gz id mol type mass x y z xu yu zu vx vy vz q
fix 2 all nve/limit 0.1
fix 3 all temp/rescale 100 1.0 1.0 1.0 0.2
run 150000
write_data data*
write_restart restart.equil