Dear All,
I am seeking help for the following problem. I am using lj/cut/coul/long 12 morse. for Leonard jones and morse potential (Au-S). I am getting error: illegal pair_style. Please help me sort out this problem. Thank you in advance.
Hi,
It will be impossible to help you if you don’t provide the exact command you wrote in the input.
Simon
Thank you for your prompt response. Following is the input i am using.
units real
dimension 3
atom_style full
pair_style hybrid lj/cut/coul/long 12 morse
pair_modify tail yes
bond_style harmonic
angle_style harmonic
dihedral_style opls
kspace_style pppm 0.00001
special_bonds lj/coul 0.0 0.0 0.5
boundary p p f
read_data lammps.data
read_restart r1_min.restart
include params.sam
neighbor 2.0 bin
neigh_modify delay 10
timestep 2.0
run_style verlet
group sam type <= 56
group gold type 57
velocity sam create 300.0 23482341 mom yes rot yes dist gaussian
velocity gold create 0.0 23482341 mom yes rot yes dist gaussian
fix 1 gold setforce 0.0 0.0 0.0
#fix 2 sam setforce NULL NULL 0.0
fix 3 all nvt temp 300.0 300.0 500.0
#fix_modify 2 energy yes
thermo_style multi
thermo 500
thermo_modify flush yes
dump 1 all dcd 10000 poly.dcd
dump_modify 1 unwrap yes flush yes
restart 5000 poly_A.rest poly_B.rest
#minimize 1.0e-6 1.0e-8 10000 20000
run 500000
write_restart rest.poly
The morse sub-style requires a cutoff as argument. That argument is missing here.
pair_style hybrid lj/cut/coul/long 12 morse 2.5. This pair style also give error.
ERROR: Expected floating point parameter instead of ‘lj/cut/coul/long’ in input script or data file (src/KSPACE/pair_lj_cut_coul_long.cpp:628)
Last command: pair_coeff 1 1 lj/cut/coul/long 0.066 3.5 # C1 C1
following is how my parameters file for leonard jone and morse potential look like. Can you please have a look wether my parameters file is correct or no?
pair_coeff 1 1 lj/cut/coul/long 0.066 3.5 12 #C1C1
pair_coeff 1 2 lj/cut/coul/long 0.066 3.5 12 #C1C2
pair_coeff 57 19 morse 8.763 1.47 2.65 2.5 #Au57S19
The morse cutoff doesn’t sound realistic. Your cutoff is smaller than the minimum of the potential.
This error is only possible, if you have a pair_style lj/cut/coul/long 12 command somewhere after your pair_style hybrid command or you made some other change that you don’t show us.
As was pointed out to you earlier, it is not possible to make an assessment without seeing the whole thing.
Please also keep in mind that all we are talking about here is stuff that is described at great detail in the LAMMPS manual and that you should be able to resolve this all by yourself. All you have to do is to correctly follow the documentation and take some time to do that thoroughly and in stages.
I am getting the impression that you need more help than what can be provided in an online forum and thus should look for a local tutor.
Thank you once again. Following is all the files that i generated and using for MD simulation.
inp.lammps
units real
dimension 3
atom_style full
pair_style hybrid lj/cut/coul/long 12 morse 2.5
pair_modify tail yes
bond_style harmonic
angle_style harmonic
dihedral_style opls
kspace_style pppm 0.00001
special_bonds lj/coul 0.0 0.0 0.5
boundary p p f
read_data lammps.data
include params.sam
neighbor 2.0 bin
neigh_modify delay 10
timestep 2.0
run_style verlet
group sam type <= 56
group gold type 57
velocity sam create 300.0 23482341 mom yes rot yes dist gaussian
velocity gold create 0.0 23482341 mom yes rot yes dist gaussian
fix 1 gold setforce 0.0 0.0 0.0
fix 3 all nvt temp 300.0 300.0 500.0
thermo_style multi
thermo 500
thermo_modify flush yes
dump 1 all dcd 10000 poly.dcd
dump_modify 1 unwrap yes flush yes
restart 5000 poly_A.rest poly_B.rest
run 500000
write_restart rest.poly
parameters file
params.sam
#Masses
mass 1 12.011
mass 2 12.011
mass 3 12.011
mass 4 12.011
mass 5 12.011
mass 6 12.011
mass 7 12.011
mass 8 12.011
mass 9 12.011
mass 10 12.011
mass 11 12.011
mass 12 12.011
mass 13 12.011
mass 14 12.011
mass 15 12.011
mass 16 12.011
mass 17 12.011
mass 18 12.011
mass 19 32.06
mass 20 1.008
mass 21 1.008
mass 22 1.008
mass 23 1.008
mass 24 1.008
mass 25 1.008
mass 26 1.008
mass 27 1.008
mass 28 1.008
mass 29 1.008
mass 30 1.008
mass 31 1.008
mass 32 1.008
mass 33 1.008
mass 34 1.008
mass 35 1.008
mass 36 1.008
mass 37 1.008
mass 38 1.008
mass 39 1.008
mass 40 1.008
mass 41 1.008
mass 42 1.008
mass 43 1.008
mass 44 1.008
mass 45 1.008
mass 46 1.008
mass 47 1.008
mass 48 1.008
mass 49 1.008
mass 50 1.008
mass 51 1.008
mass 52 1.008
mass 53 1.008
mass 54 1.008
mass 55 1.008
mass 56 1.008
mass 57 64.388370
Sorry but I am unable to upload the files with the option given upload in the menu. It’s not allowed for new users. Any other possible way to send the files? Thank you in advance.
There are no settings for pair coefficients anywhere here. Where are they?
Nobody can help you, if you don’t provide consistent and complete information.
My patience is an an end.
params.sam (109.3 KB)
inp.lammps.rtf (1.3 KB)
These files are a bit difficult to read… what text formatting are you using for the input file? Most importantly the “data.lammps” is missing so it is impossible to test them.
inp.lammps.lammps (974 Bytes)
lammps.data.data (7.8 KB)
params.sam.sam (109.7 KB)
When I run your script using the 2Aug2023 version of LAMMPS, I get a different error:
ERROR on proc 0: Out of range atoms - cannot compute PPPM (…/pppm.cpp:1839)
No surprise here. The input deck is full of mistakes.
- I already mentioned the bogus cutoff of the Morse potential
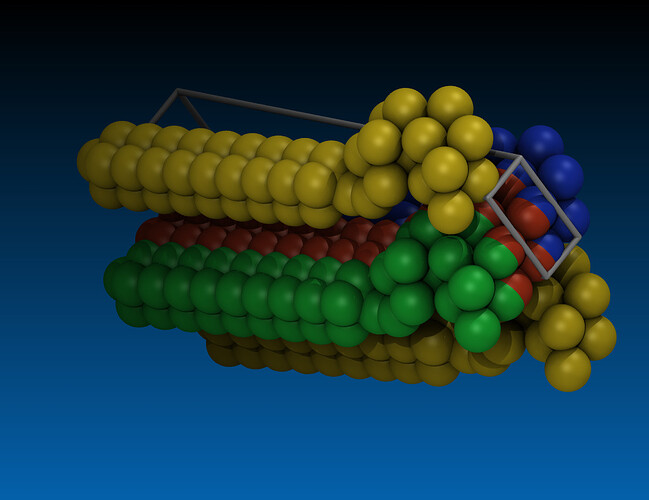
- The box dimensions are bogus and thus periodic images overlap (in x) or have a gap (in y) or the gap is (in z) not present (but it should be big). See the attached visualization where the red atoms are the original box and the colored ones periodic replica
- There are bad settings: timestep is too large, bogus neighborlist settings, kspace force error far too large
- There should be some kind of box relaxation in x and y. But then again, the proper box dimensions in x and y should be known beforehand and the potential used for the material is not very accurate and those atoms are kept immobile anyway.
- There seem to be incorrect assignments of angles or dihedrals
This whole input looks like it was done by somebody with limited understanding and no checking of whether inputs are correct or meaningful.
As mentioned before, this is not something that can be rectified in a forum with a few “do this, not that” kind of statements, but requires proper tutoring by an experienced practitioner in MD simulations that will lead to a proper understanding of the physics of the model and methods used to represent that physics.