Generating the correct improper interactions in LAMMPS (or any other simulation software) can be a long and complicated task. I recommend choosing a molecule builder instead of trying to do this by hand.
The answer to your question:
It completely depends on which improper_style you want to use. Unlike some MD programs, LAMMPS gives you the freedom to choose the equation used to describe forces between the 4 atoms in an improper interaction. (The “improper style”. And if none of the existing improper styles use the equation you want, then it is relatively simple to create your own custom improper style that uses the equation you want by editing the LAMMPS code.) Some of the existing improper styles are listed at the end of this web page:
https://lammps.sandia.gov/doc/improper_distance.html
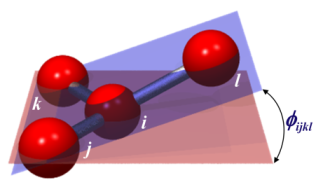
If you click on the links at the end of this page, you will see that there are many different improper styles. Some of them do not even use “angles”, like the φijkl angle that you showed here. For example, some improper interactions have equations that only depend on the height of a tetrahedron formed by the 4 atoms. For example, see:
https://lammps.sandia.gov/doc/improper_distance.html
You will have to choose which improper style you want to use (which itself is usually determined by the force field you have chosen), and then read the documentation for that improper style carefully. Different improper styles in LAMMPS use different atom order. For some improper styles, the central atom is assumed to be the first atom in the list (of atoms which appear in the “Impropers” section of your data file). For other improper styles, the central atom is the second or third atom.
Suggestion:
-First you must decide what kind of molecule you want to simulate,
-then what kind of force field you want to use,
-then look up what kind of equation that force field uses to describe the improper interactions
-then look through the list of improper styles available in LAMMPS, and pick the one which matches the force field you are using
-then look at the order that the atoms appear in that improper style (ie, which one of these 4 atoms is the central atom?).
-then create a LAMMPS data file which lists the atoms in that order.
Trying to do this by hand is usually laborious. Alternatively, I think the majority users choose a molecular dynamics program (eg LAMMPS, GROMACS, NAMD, OpenMM), which comes with a decent molecule builder which uses this force field. (Trying to build a LAMMPS data file by hand is possible. But it’s complicated enough that in most cases, I would not recommend it.)
(Welcome to MD.)
Cheers
Andrew