Hello Ovito community,
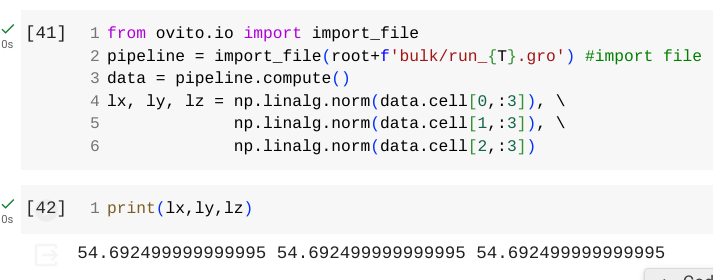
I hope this message finds you well. I am currently facing an issue while attempting to read a .gro file using Ovito’s Python scripting interface. In the process, I have observed that, upon reading the file, the comma appears to shift one position to the right, as illustrated in the attached image.
Data File (box : 5.46925 5.46925 5.46925):
Ovito output:
I’m puzzled by this unexpected behavior and would appreciate any insights or guidance on why this might be happening. Has anyone encountered a similar problem before? If so, what steps were taken to resolve it?
Your assistance in resolving this matter would be greatly appreciated. Thank you in advance for your time and expertise.
Best regards,
Martín.