Hi,
I am using fix rigid to glue beads in a rod. I relax my system to a zero force configuration using 0-temp Langevin dynamics. when I checked a pair of rigid rods with 3 beads in each rod, I found that the fix constraint is not exactly kept, and the error is in the sixth digit. the rods are slightly bending.
I put four parallel rods (short or long) in a box which are free floating in space. then I relax them and calculate sum of the bead_wise Virial_x. My hypothesis is that since the bundle is not connected to anywhere, the virial in x direction has to be zero. These are the result I get from LAMMPS: a = spacing between beads, sigma = box units, box length = 60
this is the LAMMPS input file:
atom_style molecular
read_data data_4r3B.dat
mass 1 1
mass 2 1fix fixRods all rigid molecule
fix fixLangevinOSC all langevin 0.0 0.0 10.0 1234 zero yes
fix fixNVEOSC all nve
timestep 0.05
compute PotentialEOSC all pe
fix fixPEOSC all ave/time 1 1 1 c_PotentialEOSC file pot_relaxing_eps_0.txtcompute peratomOSC all stress/atom pair
compute pOSC all reduce sum c_peratomOSC[1] c_peratomOSC[2] c_peratomOSC[3] c_peratomOSC[4] c_peratomOSC[5] c_peratomOSC[6]thermo 10
thermo_style custom step temp pe etotal press xlo xhi
thermo_modify flush yes
dump dumpCus all custom 10000000 4rods3B.xyz.* type x y z fx fy fz c_peratomOSC[1] c_peratomOSC[2] c_peratomOSC[3] vx vy vz
dump_modify dumpCus format “%d %20.15g %20.15g %20.15g %20.15g %20.15g %20.15g %20.15g %20.15g %20.15g %20.15g %20.15g %20.15g” sort idpair_style lj/cut 1.58
pair_coeff 1 1 0.001 1.0 1.1225
pair_coeff 2 2 0.10 1.0 1.58
pair_coeff 1 2 0.001 1.0 1.1225
neigh_modify exclude molecule all
log log_manualRelax_eps_0.dat
run 10000000
- 4rods, 3beads in each, a/sigma = 1.0 —> LAMMPS Virial_x = -2.9e-13 - This case LAMMPS giving zero value for virial.
beads are colored based on Virial_x
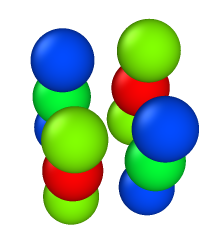
- 4rods, 21beads in each, a/sigma = 1.0 —> LAMMPS Virial_x = -5.16e-5 - LAMMPS reports non-zero virial in x direction.
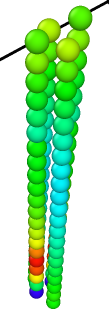
- 4rods, 51beads in each, a/sigma = 1.0 —> LAMMPS Virial_x = -0.00153210 - LAMMPS reports non-zero virial in x direction.
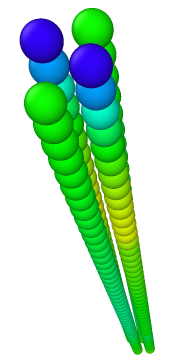
- 4rods, 51beads in each, a/sigma = 0.4 —> LAMMPS Virial_x = -0.0289228943
Looks like error depends on BOTH Number of beads in a rod, and a/sigma.
In all case studies, all the beads are type 2 particles.
Do you have any insight on why I would get non-zero virial for a free floating bundle? Any help would be appreciated
Thanks,
-Navid Kazem
PhD Candidate, Soft Machines Lab
Civil and Environmental Engineering Department
Carnegie Mellon University


