Hello All,
I have been working on developing a code for a Simple Eukarotic Cell within Lammps.
This paper ( Lennard-Jones type pair-potential method for coarse-grained lipid bilayer membrane simulations in LAMMPS - ScienceDirect) has been a major help but I am at somewhat of an impasse now.
When I run my simulation, it goes fine until i try to read the data file, once this happens I get the error message "Segmentation Fault (Core Dumped) " and the sim cancels out.
I have done some digging on this message and I haven’t seen anything directly on it, just some old posts that didn’t help too much.
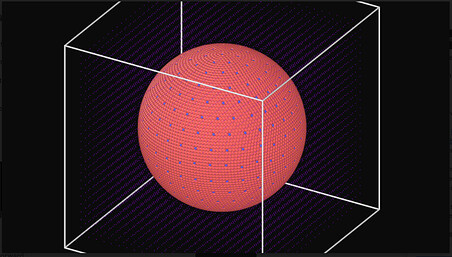
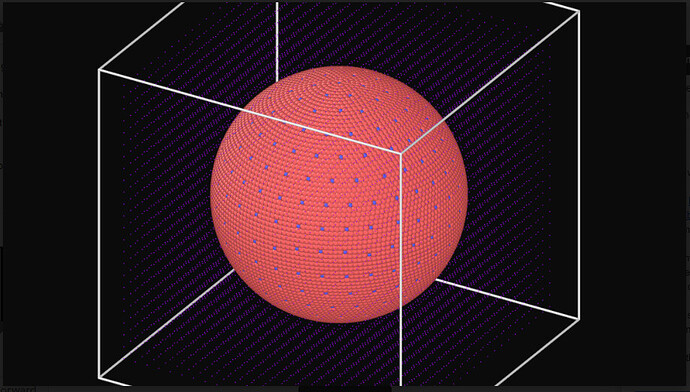
Included are the input file I am trying to run, the first few lines of the data file, and a picture of the data file ( as the full file is wayyy to long)
Input File
# Cell Simulation
variable infile string cleaned-up-v1
variable datafile string RBC_V2
variable P_damp equal 1 # Sets the Pressure damping variable used in the following " fix npt" commands
variable P equal 0.05 # Sets the desired pressure of the system used in the following " fix npt" commands
variable scale1 equal ramp(2.7,2.52) # Sets the scale performed in the final fix on water scaling down sigma from 2.7- 2.5
variable ew equal 0.2 # Sets the energy well ( epsilon) variable used in the pair coefficient specifications
variable ini_T equal 0.02 # Sets the initial temperature used in the fix commands below
variable T equal 0.23 # Sets the final temperature used in the fix commands below
variable LD equal 1.0 # Sets the temperature damping parameter used in fix commands below
variable beta equal 0.0194*0.0
units lj
atom_style hybrid ellipsoid peri molecular
bond_style harmonic # Sets the bond style as harmonic spring defined by Hooke’s law
boundary p p p # Creates the periodic boundaries of the simulation box
read_data ${datafile}.data # Specifies which read data file will be used to read in initial coordinates of system
# spectrin bonds
bond_coeff 1 50 1.5 # Defines the bond coefficients and spring constant for the spectrin network bonds
# membrane-spectrin anchoring
bond_coeff 2*3 20 2.0 # Defines the bond coefficients and spring constant for the transmembrane protein to spectrin network bonds
mass * 1.0
set type 1*2 shape 1 1 1 # membrane particle are rigid body, it needs shape parameter to be set
group bilayer type 1 2 # Creates a group called bilayer which consists of type 1 and 2 particles,
#(Type 1 Bilayer, Type 2 Transmembrane proteins)
group network type 3 4 5 # Creates a group called network which consists of type 3 4 5 particles
group anchor_bonds type 2 3 5
group water_in type 6 # Creates a group called water_in consisting of type 6 particles ( Type 6 = Water inside Membrane)
group water_out type 7 # Creates a group called water_out consisting of type 7 particles ( Type 7 = Water Outside Membrane)
group water type 6 7 # Creates a group called water consisting of both type 6 and 7 particles ( Water inside + Out)
velocity bilayer create {T} 87287 loop geom # Creates Initial Velocity for bilayer particles ( Type 1 and 2)*
*velocity network create {ini_T} 87287 loop geom # Creates Initial Velocity for network particles ( Type 3 and 4 and 5)
velocity water create ${T} 87287 loop geom # Creates Initial Velocity for water particles ( Type 6 and 7 )
pair_style hybrid lj/cut 3.6 # Sets the pair_style potentials used for the simulation,
#in the case both lennard-jones/cut and the fluidmembrane potential
pair_coeff 12 12 lj/cut {ew} 1.0 # Sets the pair coefficients and parameters between type 1 and 2 (bilayer) particles</i>
<i>pair_coeff 1*2 3*5 lj/cut {ew} 1.0 # Sets the pair coefficients (Epsilon, Sigma) and parameters between type 1,2 (bilayer) and 3,4,5 ( Network Particles)
pair_coeff 12 67 lj/cut {ew} 1.0 # Sets the pair coefficients (Epsilon, Sigma) for interaction between type 1,2 (bilayer) and 6,7 (water) particles</i>
<i>pair_coeff 3*5 3*5 lj/cut {ew} 1.0 # Sets the pair coefficients (Epsilon, Sigma) for interaction between type 3,4,5 (network)
# and type 3,4,5 (network) particles
pair_coeff 35 67 lj/cut ${ew} 1.0 # Sets the pair coefficients (Epsilon, Sigma) for interaction between type 3,4,5 (network)
# and type 6,7 (water) particles
pair_coeff 67 67 lj/cut ${ew} 2.7 # Sets the pair coefficients (Epsilon, Sigma) for interaction between type 6,7 (water)
# and type 6,7 (water) particles
neighbor 1.0 bin
thermo_style custom step temp press ebond # Printing thermodynamic data to the screen and log file
thermo 200 # Sets the number of timesteps to run before printing out thermodynamic data
timestep 0.005 # Sets the timestep of the simulation
### equilibrium
fix 1 water npt temp {T} {T} {LD} iso {P} {P} {LD}
dump 1 all atom 500 dump.equili.${infile)
run 10000
fix 2 network npt temp {ini_T} {T} {LD} iso {P} {P} {P_damp}
undump 1
dump 2 all atom 500 dump.run.${infile).#1
run 10000
fix 3 bilayer nvt/asphere temp {T} {T} {LD}*
*undump 2*
*dump 3 all atom 500 dump.run.{infile).#2
run 10000
### change RBC volume
fix 4 water adapt 1 pair lj/cut sigma 6 6 v_scale1
undump 3
dump 4 all atom 500 dump.run.${infile).#3
run 10000
unfix 4
undump 4
dump 5 all atom 500 dump.run.${infile).#4
run 10000
Data File
#RBC_V2.data
21702 atoms
3202 bonds
8346 ellipsoids
7 atom types
3 bond types
-35 35 xlo xhi
-35 35 ylo yhi
-35 35 zlo zhi
Atoms
1 2 0.004 0.005 25.002 1 0.239 0.524 0
2 2 0.000 0.002 -24.998 1 0.239 0.524 0
3 1 0.971 0.009 24.991 1 0.239 0.524 0
4 1 0.488 0.840 24.982 1 0.239 0.524 0
5 1 -0.476 0.840 24.989 1 0.239 0.524 0
6 1 -0.961 0.006 24.991 1 0.239 0.524 0
7 1 -0.484 -0.830 24.982 1 0.239 0.524 0
8 1 0.494 -0.832 24.987 1 0.239 0.524 0
9 1 1.947 0.007 24.935 1 0.239 0.524 0
10 1 1.724 0.904 24.933 1 0.239 0.524 0
11 1 1.106 1.601 24.930 1 0.239 0.524 0
12 1 0.243 1.927 24.933 1 0.239 0.524 0
13 1 -0.677 1.816 24.935 1 0.239 0.524 0
14 1 -1.444 1.294 24.928 1 0.239 0.524 0
15 1 -1.873 0.464 24.929 1 0.239 0.524 0
16 1 -1.879 -0.458 24.933 1 0.239 0.524 0
17 1 -1.445 -1.276 24.932 1 0.239 0.524 0
18 1 -0.681 -1.807 24.926 1 0.239 0.524 0
19 1 0.240 -1.919 24.926 1 0.239 0.524 0
20 1 1.101 -1.591 24.925 1
Picture of Data
Thanks so much for any help you can provide,
Jack