Hello,
(My LAMMPS version is from 29 Oct 2020 (a little out of date but don’t think the issue is there), and the pizza.py I’m running is the installed through conda, it appears the last change was March 23 2014).
I’m trying to visualize a chain polymer I’m simulating; the topology itself is quite simple (bead n is connected n-1 and n+1, except for the ends, which have just one bond). My simulation runs a hybrid angle ellipsoid atom_style, and my initial attempts at visualizing come from lammpstrj from dump commands like the following:
dump 25 all custom 250 mdmains.lammpstrj id type x y z
(I’ve attached an example lammpstrj file)
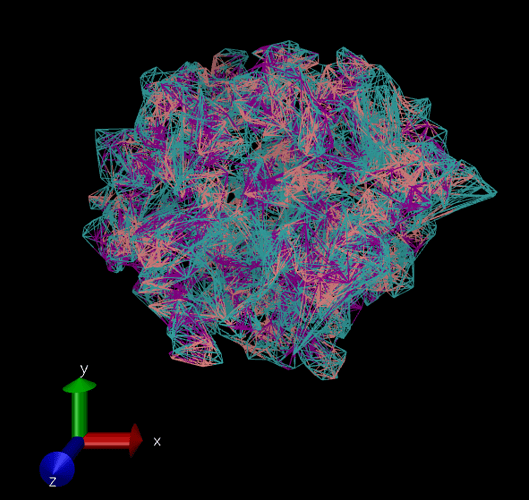
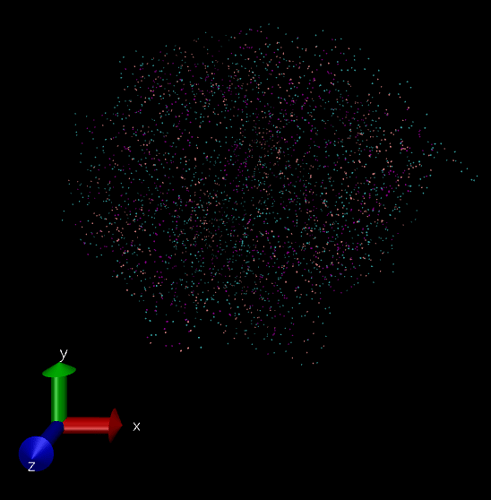
The issue being when I load it into VMD, the bond info isn’t there. So it tries to ‘infer’ the bonds (I think), and it’s a mess (image attached), so I’ve been just using the ‘Points’ drawing method (under Graphical Representations) (image attached). However, I’d like to visualize with the actual bonds, to show a chain polymer. My thought then was to generate a “molecule” file (with bond info), then I could put in a trajectory file with the “molecule” file to get the 3D coordinates, and their changes in time. This is a fairly basic VMD usage pattern, if I remember correctly.
I thought I could potentially utilize the pizza.py package, although I’ve run into a couple of dead ends, and probably am misunderstanding something. First, I generate a bond dump file with the following command:
compute bA all property/local btype batom1 batom2
dump bAAA all local 1000 tmp.dump index c_bA[1] c_bA[2] c_bA[3]
(I also need to edit the resulting dump file to match the format for bdump in the manual). Then in pizza.py I run
b = bdump(“tmp.dump”)
p = pdbfile(b)
But if I try to export as pdb file with p.one(), I get the following:
“AttributeError: bdump instance has no attribute ‘iterator’”
Looking at the chart at the bottom of this page, it seems that ‘dump.py’ and ‘bdump.py’ are not interchangeable. How then should I get a molecule file (like a pdb file) that has the correct bond information?
I also thought that perhaps I could get the info from the data file. It doesn’t seem to work with ellipsoid files, so first I edited my data file and got rid of the ellipsoid parts; with that change, I was able to load in the datafile with (and then convert to a pdbfile object):
d = data(“data.file”)
p = pdbfile(d)
However, again, if I try to run p.one() (or some other save method), it gives me an error, this time the following:
line 292, in viz
id = self.names[“id”]
KeyError: ‘id’
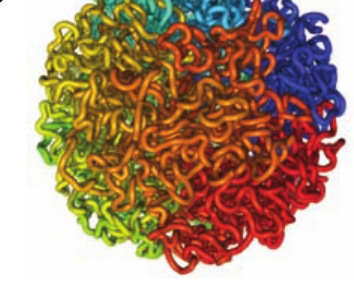
Feeling quite stuck in the pizza.py manual at this point, and wondering any pointers on how to get this visualization (I’ve attached a goal picture as ‘goal.png’, from lieberman-aiden 2009).
Best,
Christian