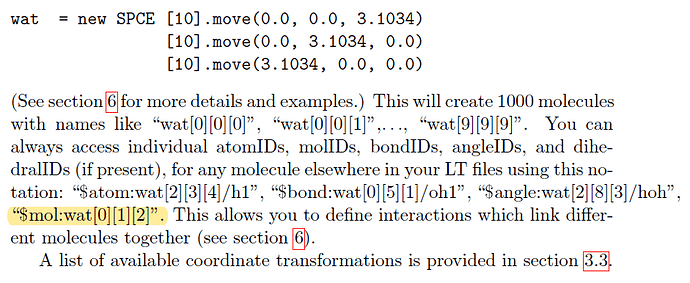
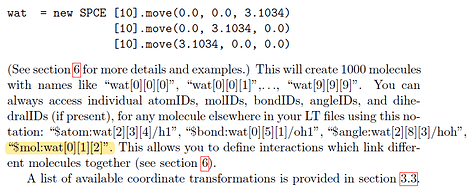
Good weekend, moletemplate developers and users, I want to inquire about that,
One lipid molecule conisist of several atom had been built with command:
Then, I copy the lipid to define a lipid membrane domain with:

The double-layer rectangular domain is created. The first question is how to know the rotated lipid molecule id because they are changed after rotation. before rotation the molecule id on the four edges are:
$mol:lipids[][0][0] $mol:lipids[0][][0] $mol:lipids[13][][0] $mol:lipids[][13][0]
as the manual told me,
Now I want to group the molecule on the four edges of the domain and use the command “group”
But the lammps doesn’t recognize the molecule id form.
I’ll start with this question first, because I think it is
the source of your misunderstanding.
This is a current limitation of moltemplate. This is not a LAMMPS issue.
In moltemplate, you can use wildcards in arrays when you want to
delete or .move() (or rotate) multiple entries in the array at the same time.
You can also use wildcards with @atom type names in pair_coeff,
commands, and also in “Data Bonds by Type” “Data Angles By Type”,
“Data Dihedrals By Type”, and “Data Impropers By Type” sections.
But you cannot use * in a LAMMPS “group” command.
I don’t blame you for trying to do this. It’s a good idea. But this notation
does not work. If you’re curious what will happen see the end of this email.
When I wrote moltemplate, it never occurred to me that somebody would
attempt to define a group this way. In retrospect, it makes sense that
someone would try this.
I’m embarrassed to admit that moltemplate does not have for-loops either.
So currently, if you want to define a group of lipids at the edge of the array,
you must use a long tedious list of commands, adding each lipid,
one lipid at a time:
Add lipids[i][j][k] to group “BC” for “i”=0
I assume you want the lipids at the top and bottom
(k=0 or k=1) to be included:
group BC molecule $mol:lipids[0][0][0]
group BC molecule $mol:lipids[0][0][1]
group BC molecule $mol:lipids[0][1][0]
group BC molecule $mol:lipids[0][1][1]
group BC molecule $mol:lipids[0][2][0]
group BC molecule $mol:lipids[0][2][1]
:
group BC molecule $mol:lipids[0][12][0]
group BC molecule $mol:lipids[0][12][1]
Add lipids[i][j][k] to group “BC” for “i”=12
(k=0 or k=1) to be included:
group BC molecule $mol:lipids[12][0][0]
group BC molecule $mol:lipids[12][0][1]
group BC molecule $mol:lipids[12][1][0]
group BC molecule $mol:lipids[12][1][1]
group BC molecule $mol:lipids[12][2][0]
group BC molecule $mol:lipids[12][2][1]
:
group BC molecule $mol:lipids[12][12][0]
group BC molecule $mol:lipids[12][12][1]
Add lipids[i][j][k] to group “BC” for “j”=0
This assumes the array is a 13 x 13 x 2 array
group BC molecule $mol:lipids[0][0][0]
group BC molecule $mol:lipids[0][0][1]
group BC molecule $mol:lipids[1][0][0]
group BC molecule $mol:lipids[1][0][1]
group BC molecule $mol:lipids[2][0][0]
group BC molecule $mol:lipids[2][0][1]
:
group BC molecule $mol:lipids[12][0][0]
group BC molecule $mol:lipids[12][0][1]
Add lipids[i][j][k] to group “BC” for “j”=12
group BC molecule $mol:lipids[0][12][0]
group BC molecule $mol:lipids[0][12][1]
group BC molecule $mol:lipids[1][12][0]
group BC molecule $mol:lipids[1][12][1]
group BC molecule $mol:lipids[2][12][0]
group BC molecule $mol:lipids[2][12][1]
:
group BC molecule $mol:lipids[12][12][0]
group BC molecule $mol:lipids[12][12][1]
I might modify Chapter 6 of the moltemplate manual to clarify this limitation.
Meanwhile, I’ll treat this as a suggestion for the future.
If I have time, perhaps I will implement the ability to use [*] in a “group” command.
(Incidentally, if you are using the MARTINI lipid model, please email me because I have some comments that might be useful.)
Andrew
PS. Moltemplate is not smart enough to attempt to understand LAMMPS “group”
commands. If you are curious what will happen, I think moltemplate will interpret the
“$mol:lipid[*][0][0]” literally, interpreting this as a new $mol counter variable
named “lipid[*][0][0]”, and assigning a new number to it which does not correspond
to any molecules in your simulation. (The “output_ttree/ttree_assignments.txt”
will probably contain a line “/$mol:lipid[*][0][0] 339”, or something similar, and
your simulation only has 338 lipid molecules.)