Greetings,
I’m working on a polymeric material (described by pcff) and would like to evaluate its modulus by monitoring the stress-strain behavior. The issue is that stress doesn’t show any meaningful trend and only oscillations are observed around the set overall pressure. Below are the key command lines in my input script:
fix 1 all npt temp 300 300 100 y 0 0 1000 z 0 0 1000" #strain along x
fix 2 all deform 1 x erate ${strain_rate_fs}" #where strain rate is 1e-8 1/fs
compute p all pressure thermo_temp
variable stx equal -0.101325*c_p[1] #in MPa
Despite time-averaging of v_stx, it doesn’t seem to be increasing in the course of simulation (2ns with timestep of 0.5 or 1 fs).
It would be highly appreciated if you could advise what I’m doing wrong here, or if there are better ways to approach this (for instance change_box).
Thanks in advance for your inputs.
Regards,
Monir
1 Like
The first step in cases where you don’t get the expected result is always to visualize the trajectory and see if you notice any unexpected behavior.
You also need to take into account that “soft” materials can have a very different behavior than, e.g. metals, so that significant trends may need larger systems and longer simulations and more averaging.
Have you compared your system size and simulation settings with similar studies of similar materials?
axel.
Is your model periodic in X-direction? ‘Fix deform’ won’t work if it isn’t periodic in the corresponding direction.
Sanjib
Many thanks for all your inputs.
The model is periodic in the strain direction (x). And the issue was resolved when I coupled fix deform with fix nvt/nve. As per Eliar’s suggestion, I also made the other two dimensions non-periodic through the deformation, but not sure how extensively it could affect the results.
My system size is comparable to those used in similar studies (500 molecules), however I believe results could be further improved by increasing the chain length. Will try that in my next attempt.
Best regards,
Monir
Greetings,
Thanks again for your comments on the question above. To evaluate the young modulus (at a temperature well below Tg) I coupled “fix deform” with “fix nvt”, however, I’m running into the following issues:
-
Although within an nvt ensemble the box dimensions in y and z direction are expected to shrink, they remain unchanged through the run (for “p p p” or “p s s” boundary conditions).
-
With a low strain rate (10^-7 1/fs), there are large fluctuations in stress; while for the higher rate (10^-6 1/fs) the obtained curve is quite smooth, but deviation of the slope from the expected value would be larger.
My input script and stress vs strain graph are included below. I’d be very grateful if you could kindly share your insights on the matter and point me to my mistakes.
Thanks in advance.
Best regards,
Monir
** Initialization **
log 20P.dat
units real
atom_style full
boundary p p p
bond_style harmonic
angle_style harmonic
dihedral_style charmm
special_bonds charmm
pair_style lj/cut/coul/long 12
kspace_style pppm 1.0e-4
pair_modify mix arithmetic
** Atom/Molecule Definition **
read_restart Restart.5000000
** Setting **
neighbor 1.0 bin
neigh_modify every 1 delay 0 check yes
timestep 0.5
compute peratom all stress/atom NULL #Units are pressure*volume
compute fy all reduce sum c_peratom[1] c_peratom[2] c_peratom[3] c_peratom[4] c_peratom[5] c_peratom[6]
compute p all reduce sum c_peratom[1] c_peratom[2] c_peratom[3]
variable L equal 118.011
variable pressx equal c_fy[1]/vol101.31e-6
variable pressy equal c_fy[2]/vol101.31e-6
variable pressz equal c_fy[3]/vol101.31e-6
variable totalp equal -(c_p[1]+c_p[2]+c_p[3])/(3*vol)101.31e-6
variable strainx equal (lx-v_L)/v_L
variable strainy equal (ly-v_L)/v_L
variable strainz equal (lz-v_L)/v_L
variable tmp equal temp
fix 2 all nvt temp 250 250 100
fix 3 all deform 5000 x erate 1e-7 remap x
fix def2 all print 20000 “{pressx} {pressy} {pressz} {totalp} {strainx} {strainy} {strainz} {tmp}” file def1.txt screen no
dump Deform all atom 5000 deform.atom
dump 1 all custom 50000 3merYM.* id x y z
dump 2 all xyz 100000 3merYM.xyz
thermo 1000
thermo_style custom step temp press pe cella cellb cellc
restart 200000 Restart.*
** Run **
run 8000000 upto
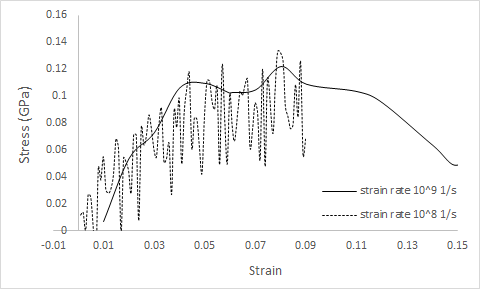
Using NVT will prevent you from observing Poisson contraction/expansion in the lateral directions. Also, I’m not sure you’ll get very good data with strain rates as low as 1e-7, but I don’t have a lot of experience with really slow strain rates in MD. My typical strain rate is 2x10^+8 1/s. Below is my usual uniaxial deformation script for polymers. Replace the force field specific section with your force field keywords.

