TLDR:
Is there a way to create a copy of the lammps C++ or Python Object? Can we deepcopy a Lammpy Object in Python (or is there a copy constructor in C++ bindings)
If not, can we reset the system back to a previous state without writing to file, e.g. by setting the positions and velocities through the Python interface?
More details
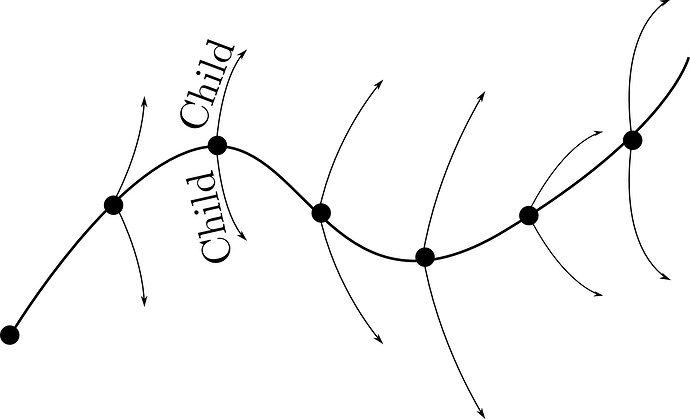
We are trying to implement a method using the Lammps Python interface which takes multil child trajectory off of a parent trajectory at intervals. The parent and child systems are identical in terms of positions, velocities, bonds, etc at the start (a mapping and force is then applied to the child). Here is an image that show the idea:
As this should run in parallel on a supercomputer (mpi4py) with lots of short runs, ideally we want to avoid writing to disk if possible. The most elegant way to do this would be to create a single parent object, evolve it in time and then copy/clone to make children runs at given intervals, run these child systems to get output and deallocate them, before running the parent forward again. Is this possible? I get an error with copy.deepcopy (which I wouldn’t expect to work for a wrapped C code anyway) and looking into the Lammps library.cpp C++ source code it seems there is no copy constructor (I assume this is not trivial to create for a general Lammps run)?
If not, is there a way to save the state of the system, e.g. the following psudo-code is a minimal example of what I want to do,
#Get the array of initial positions and velocity
nlmp = lmp.numpy
ids0 = np.copy(lmp.extract_atom("id"))
r0 = np.copy(lmp.extract_atom("x"))
v0 = np.copy(lmp.extract_atom("v"))
#Run a simulation forward
lmp.command("run 1000")
#Get pointers to variables
idsp = lmp.extract_atom("id")
rp = lmp.extract_atom("x")
vp = lmp.extract_atom("v")
#Set variables back to original values
for n in range(ids0.shape[0]):
for ixyz in range(3):
rp[idsp[n]][ixyz] = r0[ids0[n],ixyz]
vp[idsp[n]][ixyz] = v0[ids0[n],ixyz]
#This run should then be identical to the one above
lmp.command("run 1000")
When I try this, I get either errors or different output for the two runs (depending on indexing choices). Do additional variables need to be reset here or is there are better way of doing this?
I’m aware that another option to acheive the above would be,
#Get the array of initial positions and velocity
#Run a simulation forward
lmp.command("write_data file.dat ")
lmp.command("run 1000")
#This run should then be identical to the one above
lmp.command("read_data file.out")
lmp.command("run 1000")
but ideally we want to avoid the overhead of writing to disk, especially for big systems. Another option here is if write_data could be piped to a string variable or stdout.