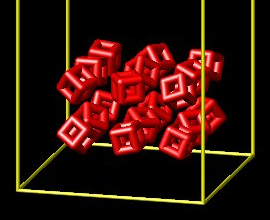
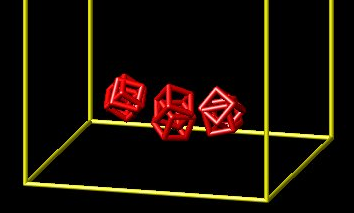
Hello, I am new to LAMMPS and as a starting point I have adapted one of the installed examples. I started with the example: mylammps\examples\body\in.cubes
My goal is to create a stack of cubes within a box, and then release one side of the box. I have been able to create the stack of cubes and save a restart file. My second input file runs for a few 2,000 time steps and then fails with the following error:
*** stack smashing detected ***: terminated
*** Process received signal ***
Signal: Aborted (6)
Signal code: (-6)
*** Process received signal ***
Signal: Segmentation fault (11)
Signal code: (128)
Failing at address: (nil)
Segmentation fault
Any help or advice would be greatly appreciated.
To create the stack of cubes, I have used this input file:
# pouring 3d rounded polyhedron bodies
variable steps index 10000
variable steps_1 index 50000
units lj
boundary fm fm fm
comm_modify vel yes
atom_style body rounded/polyhedron 1 8
atom_modify map array
region reg block 0 25 0 50 0 50 units box
create_box 4 reg
variable cut_inner equal 0.5
variable k_n equal 100
variable k_na equal 5
variable c_n equal 0
variable c_t equal 0
variable mu equal 0
variable A_ua equal 1
pair_style body/rounded/polyhedron ${c_n} ${c_t} ${mu} ${A_ua} ${cut_inner}
pair_coeff * * ${k_n} ${k_na}
neighbor 0.5 bin
neigh_modify every 1 delay 0 check yes
timestep 0.001
fix 1 all nve/body
fix 2 all gravity 1.0 spherical 0.0 -180.0
molecule object molecule.cube
#molecule object molecule.cube molecule.tetra toff 1 &
# molecule.rod3d toff 2 molecule.point3d toff 3
region slab block 5 20 30 45 25 45 units box
fix ins all pour 20 0 4767548 vol 0.4 10 region slab mol object
#fix ins all pour 600 0 4767548 vol 0.4 10 region slab mol object &
# molfrac 0.25 0.25 0.25 0.25
fix 4 all wall/body/polyhedron 2000 50 50 zplane 0.0 NULL
# Boundary
fix walls all wall/reflect xlo 0 xhi 25 ylo 25 yhi 50 zlo 0 zhi 50
#compute 1 all body/local type 1 2 3
#dump 1 all local 1000 dump.polyhedron index c_1[1] c_1[2] c_1[3] c_1[4]
#dump 10 all custom 1000 tmp.dump id type x y z radius
dump 2 all image 1000 image.*.jpg type type &
zoom 1.5 adiam 1.5 body type 0 0 view 75 15
dump_modify 2 pad 6
thermo_style custom step atoms ke pe etotal press
thermo 1000
# Restart
restart 10000 poly.restart
run ${steps}
fix ins all pour 20 0 4767548 vol 0.4 10 region slab mol object
run ${steps}
fix ins all pour 20 0 4767548 vol 0.4 10 region slab mol object
run ${steps}
fix ins all pour 20 0 4767548 vol 0.4 10 region slab mol object
run ${steps}
fix ins all pour 20 0 4767548 vol 0.4 10 region slab mol object
run ${steps_1}
fix ins all pour 20 0 4767548 vol 0.4 10 region slab mol object
run ${steps_1}
fix ins all pour 20 0 4767548 vol 0.4 10 region slab mol object
run ${steps_1}
fix ins all pour 20 0 4767548 vol 0.4 10 region slab mol object
run ${steps_1}
fix ins all pour 20 0 4767548 vol 0.4 10 region slab mol object
run ${steps_1}
fix ins all pour 20 0 4767548 vol 0.4 10 region slab mol object
run ${steps_1}
fix ins all pour 20 0 4767548 vol 0.4 10 region slab mol object
run ${steps_1}
To remove one boundary and let the particles flow out, I have used:
# Reading restart file
# Read restart file
read_restart poly.restart.390000
variable steps index 200000
#units lj
#boundary p p fm
#comm_modify vel yes
#atom_style body rounded/polyhedron 1 8
#atom_modify map array
#region reg block 0 100 0 100 0 150 units box
#create_box 4 reg
#change_box all x final 0 200 y final 0 200 z final 0 150 boundary p p f remap units box
#change_box all x final 0 100
## Variables
variable cut_inner equal 0.5
variable k_n equal 100
variable k_na equal 5
variable c_n equal 0
#20
variable c_t equal 0
#5
variable mu equal 0
variable A_ua equal 1
pair_style body/rounded/polyhedron ${c_n} ${c_t} ${mu} ${A_ua} ${cut_inner}
pair_coeff * * ${k_n} ${k_na}
neighbor 0.5 bin
neigh_modify every 1 delay 0 check yes
timestep 0.001
fix 1 all nve/body
fix 2 all gravity 10.0 spherical 0.0 -180.0
# Boundary
fix walls all wall/reflect xlo 0 xhi 25 ylo 0 yhi 50 zlo 0 zhi 50
# Bottom Boundary
fix 4 all wall/body/polyhedron 2000 50 50 zplane 0.0 NULL
thermo_style custom step atoms ke pe etotal press
thermo 1000
# Images
dump 2 all image 1000 image.*.jpg type type &
zoom 1.5 adiam 1.5 body type 0 0 view 75 15
dump_modify 2 pad 6
# Restart
restart 10000 poly.restart
run ${steps}