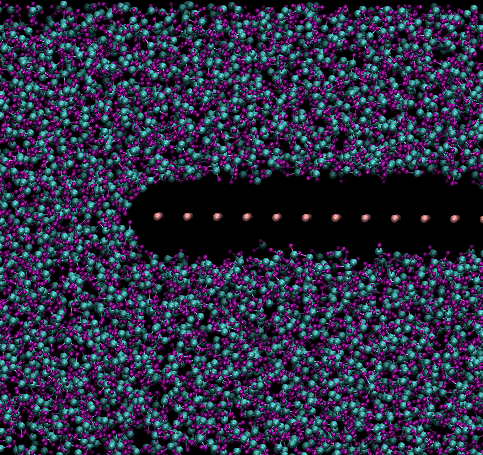
I simulated a simple sheet consisting of array of neutral atoms with
lj parameters of 0.0 0.0, to make them 'transparent'. After a while I
changed pair_coeff values to 0.0001 with sigma value of 5.0 or 5.5,
followed by structural minimization and further nvt run.
If sigma is less than 5.0, it all works as expected, however if it is
greater then 5.0, say 5.5 or 6.0, certain water molecules just freezes
where they are and do not move away (please see the picture). Is this
any known bug?
lj style : lj/cut/coul/long/gpu 10 10
LAMMPS ver: 27 August 2016 -ICMS
Run on: Xeon E5 2620 processor with 2 GTX 1080 cards
time integration fix bnvt
I simulated a simple sheet consisting of array of neutral atoms with
lj parameters of 0.0 0.0, to make them 'transparent'. After a while I
changed pair_coeff values to 0.0001 with sigma value of 5.0 or 5.5,
followed by structural minimization and further nvt run.
If sigma is less than 5.0, it all works as expected, however if it is
greater then 5.0, say 5.5 or 6.0, certain water molecules just freezes
where they are and do not move away (please see the picture). Is this
any known bug?
i don't think that there is any bug, but rather you get what you are asking for.
have you monitored the pressure?
if you keep growing your neutral atoms, you "squeeze" the rest, and
they will go wherever they find space.
keep in mind your immobile atoms correspond to a 0K area, so any
molecules/atoms close to it will be cooled down.
axel.
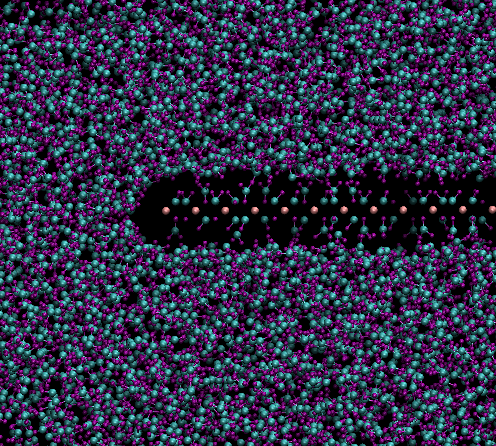
I did the simulation again with larger box size to give molecules
little room, along with few steps of simulation in beginning with lj
parameters of 4.5 for little warm up. Following this even larger lj
parameters are now working fine.
Thank you for your time

