Sorry, but this is one of the “self-explanatory” errors. If you don’t know what to do, then you don’t properly understand crucial components of your force field. It tells you that your extra/special/per/atom is too small. What do you think one would need to do in such a case?
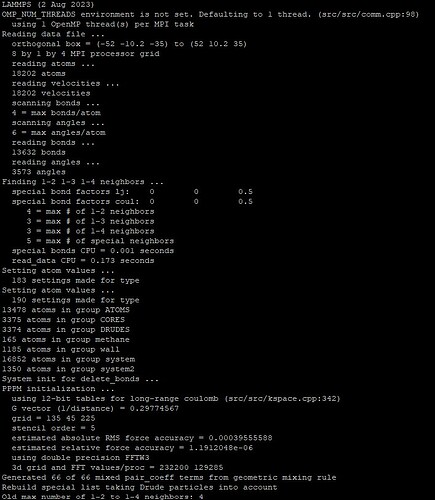
Special neighbors are atoms in the pairwise neighbor list that are either directly bonded to each other (1-2 neighbors) or bonded to a bonded atom (1-3 neighbors) or in a bond two bonds away (1-4 neighbors). These are also neighbors that would be part of bonded interactions (bonds, angles, dihedrals). In typical classical force fields, these atoms are excluded from the pair-wise neighbor list, to simplify the parameterization of bonds, angles, and dihedrals. LAMMPS maintains a list of these “special neighbors” for each atom, same as it stores bonds, angles, and dihedrals with atoms. The size of these per-atom arrays is fixed at the creation of the simulation cell, so that if the bond topology is changed later, it needs to be able to accommodate those changes. Especially, when bonds are added, the number of special neighbors (AKA exclusions) has to increase and typically by quite a bit. When all topology data is fixed, e.g. by reading a data file, LAMMPS will determine the optimum values. E.g. for the rhodo benchmark input you get:
reading atoms ...
32000 atoms
reading velocities ...
32000 velocities
scanning bonds ...
4 = max bonds/atom
scanning angles ...
8 = max angles/atom
scanning dihedrals ...
18 = max dihedrals/atom
scanning impropers ...
2 = max impropers/atom
reading bonds ...
27723 bonds
reading angles ...
40467 angles
reading dihedrals ...
56829 dihedrals
reading impropers ...
1034 impropers
4 = max # of 1-2 neighbors
12 = max # of 1-3 neighbors
24 = max # of 1-4 neighbors
26 = max # of special neighbors
The general approach when using extra/xxx/per/atom is to err on the side of being too large. While choosing too much extra property storage per atom, you increase the memory usage and communication may need to send too much data, but for the most part the impact of those are small.
Also, you can always write out a data file, when everything is assembled and its final configuration and then start from there, and it will choose the optimum values again upon reading that data file.
At any rate, how to deal with this error should be obvious now.
Probably your file is too large. Why upload such a large file in the first place. If you want people to help you, you should create small test cases, so simulations run small and debugging can be done quickly. In fact, I would content that you are making your own life harder than it needs to be when working with oversized systems while you are still trying to figure out how to set those up in the first place. Experienced people set up small tests to sort out all simulation details, before they switch to a more reasonable size for setting up a production simulation.