Dear all,
I wish this is my coding problem not a bug.
Below is the code that intends to load a full-type lammps data file and make changes to bond count, angle count, charges, then export it as charge-type lammps data. Everything seems working well, except the number of bond/angles remain unchanged though “data.particles_.bonds_.bond_types_.size” report correctly.
ps. I was supposed to update lammps data file, but a new user does not allowed to upload file…
file_pdb = "methane_oxygen_1000K.lmpdat"
output_data_file = "methane_oxygen_1000K_charge.lmpdat"
import ovito.io as ovio
pipeline = ovio.import_file(file_pdb,atom_style="full")
data = pipeline.compute()
data.particles_.bonds_.count = 0
data.particles_.angles_.count = 0
print(data.particles_.bonds_.bond_types_.size)
charges = data.particles_.get_mutable('Charge')
for i in range(len(charges)):
charges[i] = 0.0
ovio.export_file(data,output_data_file,"lammps/data",atom_style="charge")
Hi!
Let me try to explain this. If you do
data.particles_.bonds_.count = 0
you are setting the number of bonds to 0 but not the number of bond types. If you do
print(data.particles_.bonds_.bond_types_.size)
you are printing the current length of the “Bond Type” property array, which stores the per-bond type IDs (one integer value per bond). Since you have reduced the number of bonds to zero, this array also has length 0.
What you are actually interested in is the list of bond types, which is a separate list object attached to the property array:
data.particles.bonds.bond_types.types
You can clear this list as follows:
del data.particles_.bonds_.bond_types_.types[:]
This should solve your problem. And you can do the same for the angle types list.
For further information, please take a look at these doc pages:
https://docs.ovito.org/reference/pipelines/data_objects/particles.html#scene-objects-particle-types
https://docs.ovito.org/python/modules/ovito_data.html#ovito.data.Property
Hi Stukowski,
Thank you for your attention.
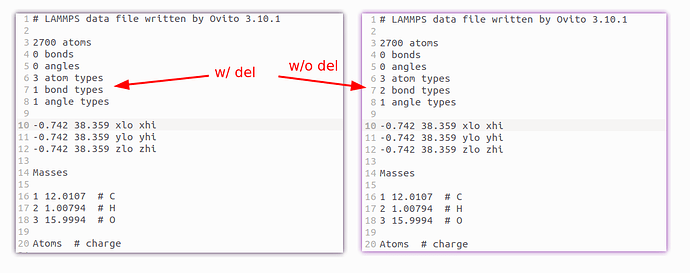
adding following to my code before executing export_file, it does help a bit. But, bond type is changed to 1, which is still not expected (supposed to be 0, as all items in the list are removed).
del data.particles_.bonds_.bond_types_.types[:]
Yijin
Yeah, right, that’s something I have overlooked so far. The LAMMPS data file exporter of OVITO implicitly defines an anonymous bond type during export if the provided bond types list is empty. That’s because the bonds in the Bonds section must have some type to yield a valid data file. They cannot be exported without having any type.
In your case, however, the Bonds section is empty, because you have 0 bonds to export. This is a special case, which the LAMMPS data exporter does not consider. Could you please provide some background on why you need to produce a data file with 0 bonds and 0 bond types? What are you going to do with it?
Hi Stukowski,
Here is the story, I am using a list of tool chain to prepare simulation box for lammps simulation, including packmol->Avogadro->ovito->lammps
Avogadro can help to export file to lammps data file(atomic style is always full), for my reaxff simulation work, I dont need bond/angles/dihedrals/impropers be defined ahead, that is why I need ovito to import_flle (with atom_style of full) then remove bond/angles/dihedrals/impropers, and finally execute export_file (with atom_style of charge) .
Btw, to get angle_types seems way different from bond_types (data.particles_.bonds_.bond_types), I cannot access angle_types in a similar way. The following code actually tells there is no angle_types attributes at all.
print(dir(data.particles_.angles_))
In fact, it seems I have to use following code to access it, am I right?
data.particles_.angles_.get_mutable("Angle Type").types[:]
Yijin
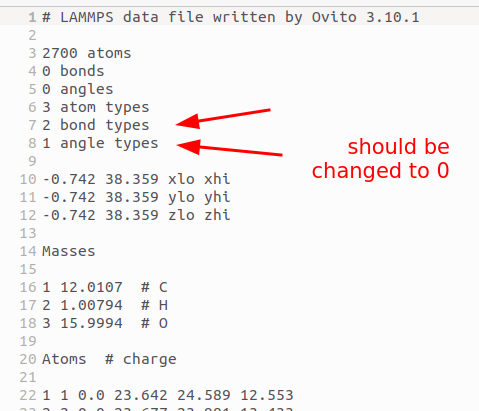
I have now made changes to the source code of OVITO so that in the next release “0 bond types” will be written to the data file if 0 bonds and 0 bond types exist in the OVITO molecule structure. The same applies to the other molecular topology types.
But there may be an even simpler solution in your case: You can completely remove the Bonds, Angles, etc. container objects from the structure:
data.particles_.bonds = None
data.particles_.angles = None
data.particles_.dihedrals = None
data.particles_.impropers = None
Then the data file exporter of OVITO won’t emit any topology-related metadata and file sections. I haven’t tested it myself, but my hope is that LAMMPS will accept such a file.
Regarding your latest question: Right, the other molecular topology containers don’t provide shortcut accessors for their type properties. You have to access these properties by real name, e.g. “Angle Type”:
del data.particles_.angles_['Angle Type_'].types[:]
The appended underscore implicitly makes the accessed property mutable. So the line above is equivalent to this full expression:
del data.particles_.angles_.make_mutable(data.particles.angles['Angle Type']).types[:]
1 Like
Hi Stukowski, it works! thanks!