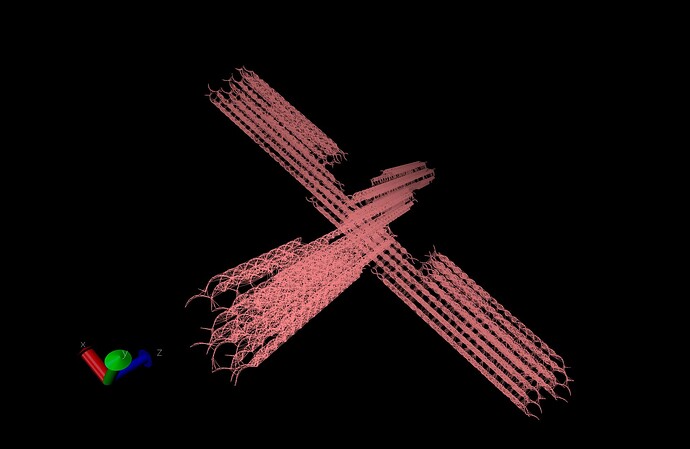
New to LAMMPS. I am using Lammps August 2 2023 version. I have run simulations with several subtle/slight variations of the same initial configuration topology, and no matter which one I use my advisor and I notice an ugly kink in the structure. So, I decided to decrease the timestep and this doesn’t seem to help the issue. I obtain my pair style and pair coefficients from the coarse grained DNA paper for oxDNA2 setups ( https://docs.lammps.org/PDF/CG-DNA.pdf ).
You can see here part of the structure seems to be moving away from the bulk of the origami. Below is a picture of the initial configuration topology, which doesn’t appear to have any of these structural inconsistencies as above:
Because the input topology has no issues with this, I can rule out that the issue is with my topology file, but instead is with my script below:
# LAMMPS Input Script for DNA Origami Simulation
log /ori24/log_ori_cg10_2.lammps
variable T equal 0.1
variable rhos equal 0.2
units lj
dimension 3
boundary p p p
atom_style hybrid bond ellipsoid oxdna
atom_modify sort 0 1.0
neighbor 1.4 bin
neigh_modify every 1 delay 0 check yes
# Read the data file
read_data /ori24/output10.dat.lammps.txt
# Set masses
#set atom * mass 3.1575
# Define nucleotide groups
group all type 1 4
# Define bond, angle, and dihedral styles (specify your coefficients)
bond_style oxdna2/fene
bond_coeff * 2.0 0.25 0.7564
pair_style hybrid/overlay oxdna2/excv oxdna2/stk oxdna2/hbond oxdna2/xstk oxdna2/coaxstk oxdna2/dh
pair_coeff * * oxdna2/excv 2.0 0.7 0.675 2.0 0.515 0.5 2.0 0.33 0.32
pair_coeff * * oxdna2/stk seqdep ${T} 1.3523 2.6717 6.0 0.4 0.9 0.32 0.6 1.3 0 0.8 0.9 0 0.95 0.9 0 0.95 2.0 0.65 2.0 0.65
pair_coeff * * oxdna2/hbond seqdep 0.0 8.0 0.4 0.75 0.34 0.7 1.5 0 0.7 1.5 0 0.7 1.5 0 0.7 0.46 3.141592653589793 0.7 4.0 1.5707963267948966 0.45 4.0 1.5707963267948966 0.45
pair_coeff 1 4 oxdna2/hbond seqdep 1.0678 8.0 0.4 0.75 0.34 0.7 1.5 0 0.7 1.5 0 0.7 1.5 0 0.7 0.46 3.141592653589793 0.7 4.0 1.5707963267948966 0.45 4.0 1.5707963267948966 0.45
pair_coeff 2 3 oxdna2/hbond seqdep 1.0678 8.0 0.4 0.75 0.34 0.7 1.5 0 0.7 1.5 0 0.7 1.5 0 0.7 0.46 3.141592653589793 0.7 4.0 1.5707963267948966 0.45 4.0 1.5707963267948966 0.45
pair_coeff * * oxdna2/xstk 47.5 0.575 0.675 0.495 0.655 2.25 0.791592653589793 0.58 1.7 1.0 0.68 1.7 1.0 0.68 1.5 0 0.65 1.7 0.875 0.68 1.7 0.875 0.68
pair_coeff * * oxdna2/coaxstk 58.5 0.4 0.6 0.22 0.58 2.0 2.891592653589793 0.65 1.3 0 0.8 0.9 0 0.95 0.9 0 0.95 40.0 3.116592653589793
pair_coeff * * oxdna2/dh ${T} ${rhos} 0.815
#Special bonds setting
special_bonds lj 0.0 1.0 1.0
#Energy Minimization settings
min_style cg
minimize 1.0e-6 1.0e-8 1000 100000
#fix cmds (time integration)
fix 1 all nve
fix 2 all langevin 0.1 0.1 10 12345
# Run settings
timestep 0.0005
thermo 100
dump out all xyz 10000 /ori24/dump10_2.lammpstrj
#dump out all xtc 10000 ori24/dump10_2.xtc
#dump_modify out unwrap yes
run 5000000
I understand the timestep may be too low. We started at timestep 0.002, then moved down to 0.001, yet the issue with the output structure in VMD still exists. Otherwise, neighbor skin is set correctly for this experiment, and as I mentioned the oxDNA2 classes with its parameters are straight from their documentation, though they model a much smaller DNA fragment which leads me to believe this could be the issue.
If anybody sees something strikingly erroneous or has dabbled in oxDNA ecosystem before, please let me know which part specifically I should be most worried about.
Thanks
output10.dat.lammps.txt (6.9 MB)