Hi Ovito-team,
I was trying to visualize a structure made with ase (atomic simulation environment) with Ovito.
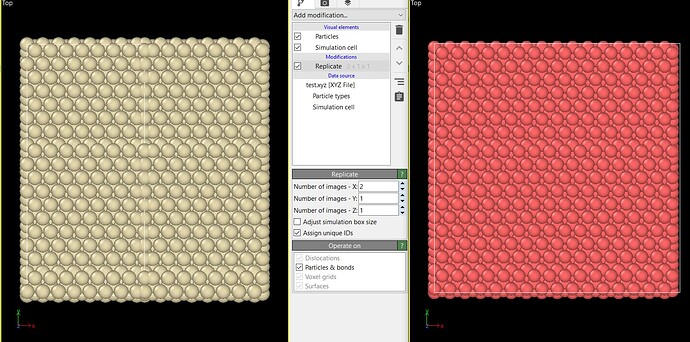
I wrote the structure to an xyz file with ase to be able to visualize it. However, when replicating the cell in x, a non-periodic structure appeared. When replicating the same structure in the python/ase tool, nothing seemed wrong. I then decided to write to a lammpsfile and visualize it with Ovito again, nothing was wrong.
Admittedly, it could be a problem with writing as well. Below, the code used to generate the structure and the xyz and lammps files:
import ase
from ase import build
from ase import visualize
from ase import io
surf = ase.build.surface(‘Pt’, (5,5,4), 20, 40, periodic = True)
surf = ase.build.make_supercell(surf, [[1,1,0],[0,2,0],[0,0,1]], wrap=True, tol=1e-05)
surf = ase.build.make_supercell(surf, [[1,0,0],[4,1,0],[0,0,1]], wrap=True, tol=1e-05)
surf.rotate(surf.cell[0], (1,0,0) , rotate_cell=True)
ase.io.write(‘test.xyz’, surf, format=‘xyz’)
ase.io.write(‘test.lammps’, surf, format=‘lammps-data’, atom_style=‘full’)
ase.visualize.view(surf)
See the image below to see what I mean with “non-periodic”. On the left, the xyz file, on the right the lammpsfile.
I solved the problem for myself, by just writing it to another filetype, but I thought it might be nice to report it as a bug.
Kind regards,
Justina