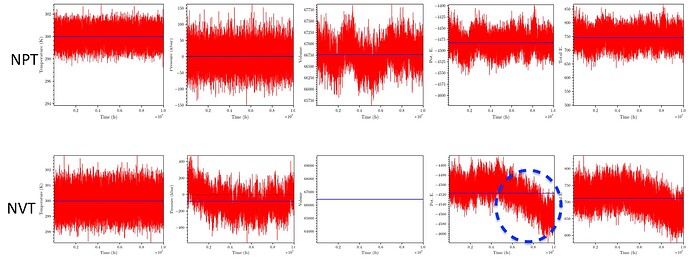
Hello, I am trying to equilibrate a system containing around 6000 atoms (a fluorinated polymer + lithium salt) using LAMMPS (23 Jun 2022 - Update 4)/Linux with OPLS-aa force-field. The OPLS parameters were obtained from LigParGen server. The full simulation includes (1) energy minimisation, (2) NVT equilibration for 1 ns, (3) NPT equilibration for 10 ns and box rescalling and (4) MD run (NVT) for 10 ns. The time step is 1fs. At the end of step 3 (NPT) the average pressure is 1 KBar, and the PE and Total E fluctuates around a mean value. However in the last NVT step, the PE and Total E starts decreasing after 5 ns (figure attached).
Also the average pressure drops substantially from 1 KBar to -80 KBar.
The input is as follows. Can someone suggest, what might be the reason? or if something is terribly wrong with the input? Thank you!
# created by fftool
units real
dimension 3
boundary p p p
atom_style full
bond_style harmonic
angle_style harmonic
dihedral_style opls
special_bonds lj/coul 0.0 0.0 0.5
# remove hybrid if not necessary
pair_style hybrid lj/cut/coul/long 12.0 12.0
pair_modify mix geometric tail yes
kspace_style pppm 1.0e-6
read_data data.lmp
# remove pair style if not using hybrid
pair_coeff 1 1 lj/cut/coul/long 0.018279 2.126000 # Li Li
pair_coeff 2 2 lj/cut/coul/long 0.117789 3.500000 # Cl1 Cl1
pair_coeff 3 3 lj/cut/coul/long 0.210000 2.900000 # O02 O02
pair_coeff 4 4 lj/cut/coul/long 0.210000 2.900000 # O03 O03
pair_coeff 5 5 lj/cut/coul/long 0.210000 2.900000 # O04 O04
pair_coeff 6 6 lj/cut/coul/long 0.210000 2.900000 # O05 O05
pair_coeff 7 7 lj/cut/coul/long 0.060000 2.900000 # F00 F00
pair_coeff 8 8 lj/cut/coul/long 0.065999 3.500000 # C01 C01
pair_coeff 9 9 lj/cut/coul/long 0.065999 3.500000 # C02 C02
pair_coeff 10 10 lj/cut/coul/long 0.065999 3.500000 # C03 C03
pair_coeff 11 11 lj/cut/coul/long 0.060000 2.900000 # F04 F04
pair_coeff 12 12 lj/cut/coul/long 0.065999 3.500000 # C05 C05
pair_coeff 13 13 lj/cut/coul/long 0.030000 2.500000 # H06 H06
pair_coeff 14 14 lj/cut/coul/long 0.030000 2.500000 # H07 H07
pair_coeff 15 15 lj/cut/coul/long 0.065999 3.500000 # C08 C08
pair_coeff 16 16 lj/cut/coul/long 0.060000 2.900000 # F09 F09
pair_coeff 17 17 lj/cut/coul/long 0.060000 2.900000 # F0A F0A
pair_coeff 18 18 lj/cut/coul/long 0.065999 3.500000 # C0B C0B
pair_coeff 19 19 lj/cut/coul/long 0.030000 2.500000 # H0C H0C
pair_coeff 20 20 lj/cut/coul/long 0.030000 2.500000 # H0D H0D
pair_coeff 21 21 lj/cut/coul/long 0.065999 3.500000 # C0E C0E
pair_coeff 22 22 lj/cut/coul/long 0.060000 2.900000 # F0F F0F
pair_coeff 23 23 lj/cut/coul/long 0.060000 2.900000 # F0G F0G
pair_coeff 24 24 lj/cut/coul/long 0.065999 3.500000 # C0H C0H
pair_coeff 25 25 lj/cut/coul/long 0.030000 2.500000 # H0I H0I
pair_coeff 26 26 lj/cut/coul/long 0.030000 2.500000 # H0J H0J
pair_coeff 27 27 lj/cut/coul/long 0.065999 3.500000 # C0K C0K
pair_coeff 28 28 lj/cut/coul/long 0.060000 2.900000 # F0M F0M
pair_coeff 29 29 lj/cut/coul/long 0.060000 2.900000 # F0N F0N
pair_coeff 30 30 lj/cut/coul/long 0.065999 3.500000 # C0O C0O
pair_coeff 31 31 lj/cut/coul/long 0.030000 2.500000 # H0P H0P
pair_coeff 32 32 lj/cut/coul/long 0.030000 2.500000 # H0Q H0Q
pair_coeff 33 33 lj/cut/coul/long 0.065999 3.500000 # C0R C0R
pair_coeff 34 34 lj/cut/coul/long 0.060000 2.900000 # F0S F0S
pair_coeff 35 35 lj/cut/coul/long 0.060000 2.900000 # F0T F0T
pair_coeff 36 36 lj/cut/coul/long 0.065999 3.500000 # C0U C0U
pair_coeff 37 37 lj/cut/coul/long 0.030000 2.500000 # H0V H0V
pair_coeff 38 38 lj/cut/coul/long 0.030000 2.500000 # H0W H0W
pair_coeff 39 39 lj/cut/coul/long 0.065999 3.500000 # C0X C0X
pair_coeff 40 40 lj/cut/coul/long 0.060000 2.900000 # F0Y F0Y
pair_coeff 41 41 lj/cut/coul/long 0.060000 2.900000 # F0Z F0Z
pair_coeff 42 42 lj/cut/coul/long 0.065999 3.500000 # C10 C10
pair_coeff 43 43 lj/cut/coul/long 0.030000 2.500000 # H11 H11
pair_coeff 44 44 lj/cut/coul/long 0.030000 2.500000 # H12 H12
pair_coeff 45 45 lj/cut/coul/long 0.065999 3.500000 # C13 C13
pair_coeff 46 46 lj/cut/coul/long 0.060000 2.900000 # F14 F14
pair_coeff 47 47 lj/cut/coul/long 0.060000 2.900000 # F15 F15
pair_coeff 48 48 lj/cut/coul/long 0.030000 2.500000 # H16 H16
pair_coeff 49 49 lj/cut/coul/long 0.030000 2.500000 # H17 H17
pair_coeff 50 50 lj/cut/coul/long 0.030000 2.500000 # H18 H18
pair_coeff 51 51 lj/cut/coul/long 0.065999 3.500000 # C19 C19
pair_coeff 52 52 lj/cut/coul/long 0.030000 2.500000 # H1A H1A
pair_coeff 53 53 lj/cut/coul/long 0.030000 2.500000 # H1B H1B
pair_coeff 54 54 lj/cut/coul/long 0.065999 3.500000 # C1C C1C
pair_coeff 55 55 lj/cut/coul/long 0.060000 2.900000 # F1D F1D
pair_coeff 56 56 lj/cut/coul/long 0.060000 2.900000 # F1E F1E
pair_coeff 57 57 lj/cut/coul/long 0.065999 3.500000 # C1F C1F
pair_coeff 58 58 lj/cut/coul/long 0.030000 2.500000 # H1G H1G
pair_coeff 59 59 lj/cut/coul/long 0.030000 2.500000 # H1H H1H
pair_coeff 60 60 lj/cut/coul/long 0.065999 3.500000 # C1I C1I
pair_coeff 61 61 lj/cut/coul/long 0.060000 2.900000 # F1J F1J
pair_coeff 62 62 lj/cut/coul/long 0.060000 2.900000 # F1K F1K
pair_coeff 63 63 lj/cut/coul/long 0.065999 3.500000 # C1M C1M
pair_coeff 64 64 lj/cut/coul/long 0.030000 2.500000 # H1N H1N
pair_coeff 65 65 lj/cut/coul/long 0.030000 2.500000 # H1O H1O
pair_coeff 66 66 lj/cut/coul/long 0.065999 3.500000 # C1P C1P
pair_coeff 67 67 lj/cut/coul/long 0.060000 2.900000 # F1Q F1Q
pair_coeff 68 68 lj/cut/coul/long 0.060000 2.900000 # F1R F1R
pair_coeff 69 69 lj/cut/coul/long 0.065999 3.500000 # C1S C1S
pair_coeff 70 70 lj/cut/coul/long 0.030000 2.500000 # H1T H1T
pair_coeff 71 71 lj/cut/coul/long 0.030000 2.500000 # H1U H1U
pair_coeff 72 72 lj/cut/coul/long 0.065999 3.500000 # C1V C1V
pair_coeff 73 73 lj/cut/coul/long 0.060000 2.900000 # F1W F1W
pair_coeff 74 74 lj/cut/coul/long 0.060000 2.900000 # F1X F1X
pair_coeff 75 75 lj/cut/coul/long 0.065999 3.500000 # C1Y C1Y
pair_coeff 76 76 lj/cut/coul/long 0.030000 2.500000 # H1Z H1Z
pair_coeff 77 77 lj/cut/coul/long 0.030000 2.500000 # H20 H20
pair_coeff 78 78 lj/cut/coul/long 0.065999 3.500000 # C21 C21
pair_coeff 79 79 lj/cut/coul/long 0.060000 2.900000 # F22 F22
pair_coeff 80 80 lj/cut/coul/long 0.060000 2.900000 # F23 F23
pair_coeff 81 81 lj/cut/coul/long 0.065999 3.500000 # C24 C24
pair_coeff 82 82 lj/cut/coul/long 0.030000 2.500000 # H25 H25
pair_coeff 83 83 lj/cut/coul/long 0.030000 2.500000 # H26 H26
pair_coeff 84 84 lj/cut/coul/long 0.065999 3.500000 # C27 C27
pair_coeff 85 85 lj/cut/coul/long 0.060000 2.900000 # F28 F28
pair_coeff 86 86 lj/cut/coul/long 0.060000 2.900000 # F29 F29
pair_coeff 87 87 lj/cut/coul/long 0.065999 3.500000 # C2A C2A
pair_coeff 88 88 lj/cut/coul/long 0.030000 2.500000 # H2B H2B
pair_coeff 89 89 lj/cut/coul/long 0.030000 2.500000 # H2C H2C
pair_coeff 90 90 lj/cut/coul/long 0.065999 3.500000 # C2D C2D
pair_coeff 91 91 lj/cut/coul/long 0.060000 2.900000 # F2E F2E
pair_coeff 92 92 lj/cut/coul/long 0.060000 2.900000 # F2F F2F
pair_coeff 93 93 lj/cut/coul/long 0.065999 3.500000 # C2G C2G
pair_coeff 94 94 lj/cut/coul/long 0.030000 2.500000 # H2H H2H
pair_coeff 95 95 lj/cut/coul/long 0.030000 2.500000 # H2I H2I
pair_coeff 96 96 lj/cut/coul/long 0.065999 3.500000 # C2J C2J
pair_coeff 97 97 lj/cut/coul/long 0.060000 2.900000 # F2K F2K
pair_coeff 98 98 lj/cut/coul/long 0.060000 2.900000 # F2M F2M
pair_coeff 99 99 lj/cut/coul/long 0.065999 3.500000 # C2N C2N
pair_coeff 100 100 lj/cut/coul/long 0.060000 2.900000 # F2O F2O
pair_coeff 101 101 lj/cut/coul/long 0.060000 2.900000 # F2P F2P
pair_coeff 102 102 lj/cut/coul/long 0.065999 3.500000 # C2Q C2Q
pair_coeff 103 103 lj/cut/coul/long 0.060000 2.900000 # F2R F2R
pair_coeff 104 104 lj/cut/coul/long 0.030000 2.500000 # H2S H2S
pair_coeff 105 105 lj/cut/coul/long 0.060000 2.900000 # F2T F2T
pair_coeff 106 106 lj/cut/coul/long 0.060000 2.900000 # F2U F2U
pair_coeff 107 107 lj/cut/coul/long 0.060000 2.900000 # F2V F2V
variable prenpt_nsteps equal 1e6
variable npt_nsteps equal 1e7
variable md_nsteps equal 1e7
variable ndump equal ${md_nsteps}/200
variable Vol equal vol
variable tot_ene equal c_kin_ene+c_pot_ene
# Compute
compute temper all temp
compute press all pressure temper
compute pot_ene all pe
compute kin_ene all ke
thermo 100
thermo_style custom step time temp press vol density pe ke v_tot_ene ecoul epair ebond eangle edihed
# Optimize cell
fix 1 all box/relax iso 0.0 vmax 0.001
min_style cg
minimize 1.0e-12 1.0e-12 200000 200000
unfix 1
reset_timestep 0
print ">>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>> pre-npt starts <<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<"
neighbor 2.0 bin
neigh_modify delay 0 every 1 check yes
timestep 1
velocity all create 300 12345
thermo 1000
thermo_style custom step time temp press vol density pe ke v_tot_ene ecoul epair ebond eangle edihed
fix shake1 all shake 0.000001 20 0 b 10 11 13 14 19 20 25 26 31 32 37 38 43 44 48 49 50 55 56 61 62 67 68 73 74 79 80 85 86 91 92 101
fix 1stnvt all nvt temp 300 300 100 # Nose-Hoover
fix 1stave all ave/time 1 1000 1000 c_temper c_press v_Vol c_pot_ene c_kin_ene v_tot_ene file firstnvt.dat
run ${prenpt_nsteps} # 1.0 ns
unfix 1stave
unfix 1stnvt
unfix shake1
print ">>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>> pre-npt ends <<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<"
reset_timestep 0
print ">>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>> npt starts <<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<"
fix 2ndave all ave/time 1 1000 1000 c_temper c_press v_Vol c_pot_ene c_kin_ene v_tot_ene file secondnpt.dat
fix 2ndnpt all npt temp 300 300 100 iso 1 1 1000
fix 2ndvol all ave/time 1 1000 1000 v_Vol ave one
variable mean_Vol equal f_2ndvol
dump myDump all custom ${ndump} npt.lammpstrj id type x y z vx vy vz fx fy fz element
dump_modify myDump sort id
dump_modify myDump element Li Cl O O O O F C C C F C H H C F F C H H C F F C H H C F F C H H C F F C H H C F F C H H C F F H H H C H H C F F C H H C F F C H H C F F C H H C F F C H H C F F C H H C F F C H H C F F C H H C F F C F F C F H F F F
run ${npt_nsteps} # 10 ns
print "Average volume after NPT = ${mean_Vol}"
print "Last volume after NPT = ${Vol}"
# Calculate the scaling factors for each dimension
variable scaleX equal (v_mean_Vol/v_Vol)^(1.0/3.0)
variable scaleY equal (v_mean_Vol/v_Vol)^(1.0/3.0)
variable scaleZ equal (v_mean_Vol/v_Vol)^(1.0/3.0)
print "scaleX = ${scaleX}"
print "scaleY = ${scaleY}"
print "scaleZ = ${scaleZ}"
# Apply the scaling to fix the volume
change_box all x scale ${scaleX} y scale ${scaleY} z scale ${scaleZ} remap
print "New volume value after change_box = ${Vol}"
undump myDump
unfix 2ndvol
unfix 2ndnpt
unfix 2ndave
print ">>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>> npt ends <<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<"
reset_timestep 0
print ">>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>> MD starts <<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<"
thermo 1000
thermo_style custom step time temp press vol density pe ke v_tot_ene ecoul epair ebond eangle edihed
fix lastmd all ave/time 1 1000 1000 c_temper c_press v_Vol c_pot_ene c_kin_ene v_tot_ene file md.dat
fix lastnvt all nvt temp 300 300 100 # Nose-Hoover
dump myDump all custom ${ndump} md_run.lammpstrj id type x y z vx vy vz fx fy fz element
dump_modify myDump sort id
dump_modify myDump element Li Cl O O O O F C C C F C H H C F F C H H C F F C H H C F F C H H C F F C H H C F F C H H C F F H H H C H H C F F C H H C F F C H H C F F C H H C F F C H H C F F C H H C F F C H H C F F C H H C F F C F F C F H F F F
run ${md_nsteps} # 10 ns
undump myDump
unfix lastnvt
unfix lastmd
write_restart restartnvt.lmp
write_data data.nvt.lmp
print ">>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>> MD ends <<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<<"