Hello everyone,
I would like to calculate the mean curvature along the surface of my surface mesh generated by ovito. I wrote this function:
def compute_mean_curvature(vertices, faces):
"""
Discrete mean curvature
"""
n_vertices = len(vertices)
L = np.zeros((n_vertices, n_vertices)) # Laplace matrix
A = np.zeros(n_vertices) # Voronoi area per vertex
for face in faces:
i, j, k = face
vi, vj, vk = vertices[i], vertices[j], vertices[k]
# edge vectors
e0 = vj - vk
e1 = vk - vi
e2 = vi - vj
# compute angles
l0 = np.linalg.norm(e0)
l1 = np.linalg.norm(e1)
l2 = np.linalg.norm(e2)
angle_i = np.arccos(np.clip(np.dot(-e1, e2) / (l1 * l2), -1.0, 1.0))
angle_j = np.arccos(np.clip(np.dot(-e2, e0) / (l2 * l0), -1.0, 1.0))
angle_k = np.pi - angle_i - angle_j
cot_i = 1 / np.tan(angle_i)
cot_j = 1 / np.tan(angle_j)
cot_k = 1 / np.tan(angle_k)
# fill Laplace matrix (cotangent weights)
for a, b, cot in [(j, k, cot_i), (k, i, cot_j), (i, j, cot_k)]:
L[a, b] += cot
L[b, a] += cot
L[a, a] -= cot
L[b, b] -= cot
# Compute area for mean curvature normalization
face_area = 0.5 * np.linalg.norm(np.cross(vj - vi, vk - vi))
A[i] += face_area / 3
A[j] += face_area / 3
A[k] += face_area / 3
# Laplace-Beltrami operator approximation
mean_curv = np.zeros(n_vertices)
for i in range(n_vertices):
delta = np.sum([L[i, j] * (vertices[j] - vertices[i]) for j in range(n_vertices)], axis=0)
mean_curv[i] = 0.25 * np.linalg.norm(delta) / A[i]
return mean_curv
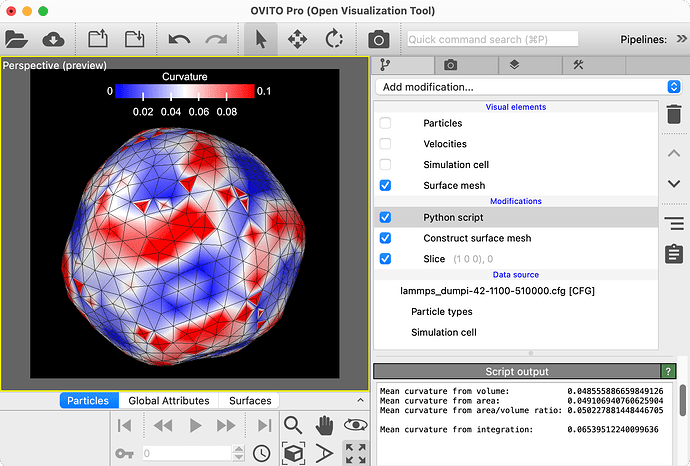
and it seems to work on a mesh that I generated using python (I made a spherical mesh and then did the numerical integration of the mean curvature along the surface which gave me approx 1/R as it should for a sphere), but once I try the same thing with the surface mesh that I generated over my spherical particle consisting of many monomers it no longer gives me 1/R. So here is my question: am I treating the mesh data wrongly and how do I have to interpret ovito’s output:
pipeline = import_file(tmp_filename)
pipeline.modifiers.append(ConstructSurfaceModifier(
method=ConstructSurfaceModifier.Method.AlphaShape,
radius=Probe_radius,
smoothing_level=Smoothing,
identify_regions=True
))
data = pipeline.compute()
mesh = data.surfaces['surface']
mesh_volume = data.attributes['ConstructSurfaceMesh.filled_volume']
mesh_surface_area = data.attributes['ConstructSurfaceMesh.surface_area']
vertex_positions_mesh = mesh.vertices["Position"]
faces_mesh = mesh.get_face_vertices()
vertices = np.array(vertex_positions_mesh, dtype=float)
faces = np.array(faces_mesh, dtype=int)
I appreciate any input, thank you in advance!
Greetings
Leah