Dear lammps user
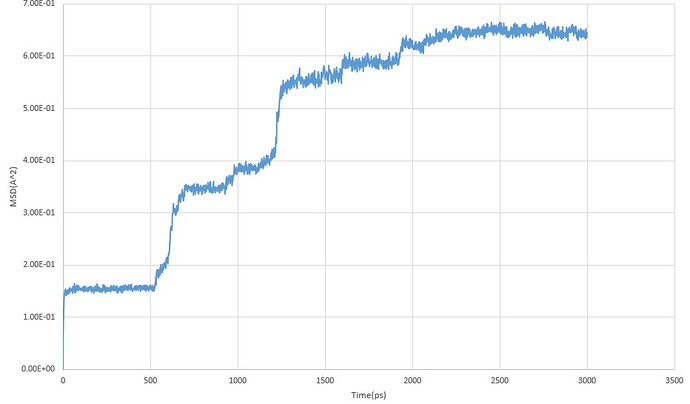
I tried to calculate diffusion coefficient thus I plotted MSD vs Time, so I used "fix Lithium_MSD Sodium ave/time 10 10 100 c_msd[1] c_msd[2] c_msd[3] c_msd[4] “.
My pair style is " lj/cut/coul/long” and the time step is 0.0001.After 30000000 run, the result was confusing me! due to this plot, could you help me to understand in which part of my work is wrong?
pair_style lj/cut/coul/long 14 14
read_data NaZr2P3O12.data
kspace_style ewald 1.0e-5
kspace_modify gewald 1.0e-5
fix MM all box/relax iso 0.0 vmax 0.001
min_style cg
minimize 1.0e-12 1.0e-12 10000 10000
unfix MM
velocity all create 700 2547321 mom yes rot yes
run 200000
unfix NPT
fix NPT all npt temp 700.0 700.1 0.01 x 0 1 1 y 0 1 1 z 0 1 1
run 30000000