Hi Everyone,
I’m facing a problem with my NPT rigid body dynamics. My system contains a TS structure in N2 solvent molecules and I’m using neural network potentials. After a few nanoseconds (15-20 ns) it fails and reports NAN for the thermodynamic data. I visualized one trajectory to see what’s happening. Here are the three snapshots before and after failing:
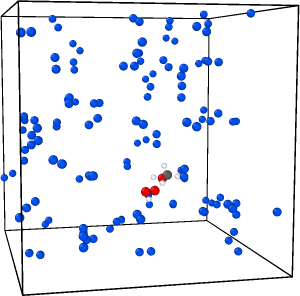
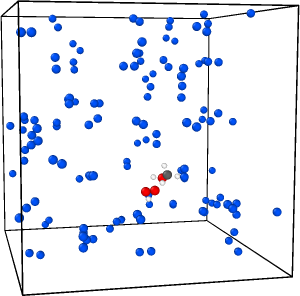
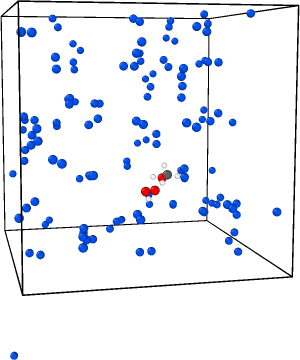
And here is the thermo data:
8651053 -198010.54 10.445166 -198000.1 750.53237 668.25858 0.22732658 13571.339
8651054 -198010.55 10.460453 -198000.09 751.63085 667.28798 0.22727077 13574.672
8651055 -198010.55 10.476788 -198000.08 752.80456 665.67571 0.22721494 13578.007
8651056 -198010.56 10.49419 -198000.06 754.055 663.40345 0.2271591 13581.345
8651057 -198010.57 10.51253 -198000.05 755.37283 660.51539 0.22710325 13584.685
8651058 -198010.57 11.087771 -197999.49 796.70644 16339.47 0.22704738 13588.028
8651059 -197974.41 13.228612 -197961.18 950.53554 15961.961 0.22698929 13591.505
8651060 -197974.45 13.28742 -197961.16 954.76121 305.85568 0.22692904 13595.114
8651061 -197974.52 13.366545 -197961.15 960.44669 283.5083 0.22686883 13598.722
8651062 -197974.61 13.471283 -197961.13 967.97258 248.24619 0.22680866 13602.329
8651063 -197974.73 13.608836 -197961.12 977.85637 198.92525 0.22674853 13605.936
And the input file:
units metal
boundary p p p
atom_style molecular
read_data “system_molecular.data”
mass 1 1.00794
mass 2 12.011
mass 3 14.0067
mass 4 15.9994
timestep 0.002
thermo_style custom step pe ke etotal temp press density vol
thermo 1
pair_style nnp dir “nnp-data”
pair_coeff * * 6.351
group RIGID molecule >= 1
fix fxlan all langevin 725 725 0.2 123456
fix fxnph RIGID rigid/nph molecule iso 500 500 2.0
run 500000
unfix fxlan
unfix fxnph
dump 1 all atom 1 dump.lammpstrj
fix fxnpt RIGID rigid/npt molecule temp 725 725 0.2 iso 500 500 2.0
run 25000000
What I’ve observed regarding this issue:
- I’m running a lot of trajectories at different pressures (from 1 bar to 500 bar) and T = 725 K for 50 ns. This problem happens randomly meaning that for some trajectories everything is fine and only a few of them fail. I use Packmol to generate the initial configurations.
- It happens more frequently at higher pressures.
- It happens less frequently for bigger boxes.
- I read here that for rigid body dynamics a small timestep is required. I tried a timestep of 1fs, but did not solve the problem.
- I also ran it in NVT, but the problem persisted.
I would appreciate it if anyone could help me with this issue. Many thanks.