hi everyone
i simulate water-copper nanofluid and use the data file for initial geometry of molecules and atoms that visualizing by vmd…
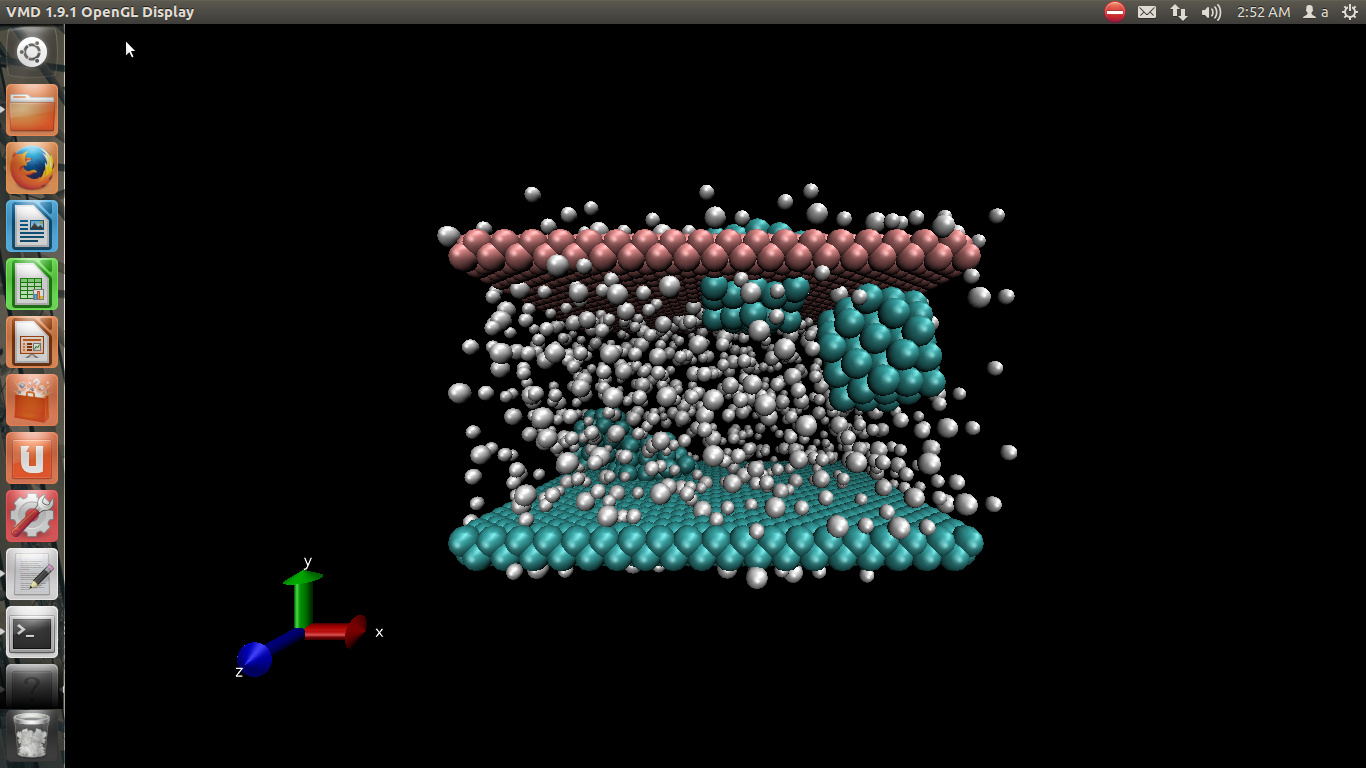
i attached picture of it in # 1.
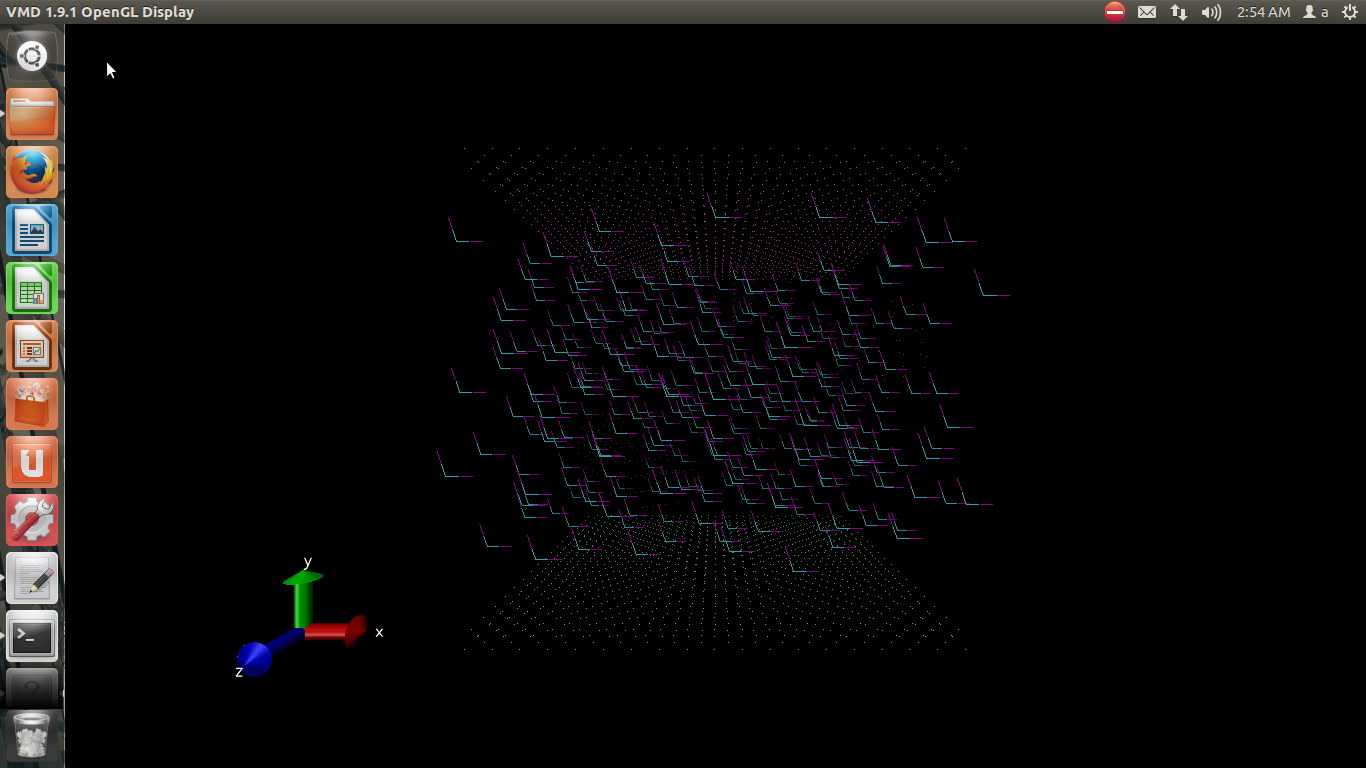
i don’t know where is my mistake beacause when i run my code with lammps the initial instruction fail and i visualize it too and attached in picture # 2.
can everyone give me some suggestions?
i modeled spc/e water molecules and use:
dimension 3
boundary p p p
atom_style full
read_data flow4s_autopsf.data
special_bonds charmm
neigh_modify delay 2 every 1
group hydrogen type 1
group oxygen type 2
group low type 3 units lattice
group up type 4 units lattice
group nanoparticle type 5 units lattice
set group hydrogen type 1
set group oxygen type 2
set group low type 3
set group up type 4
set group nanoparticle type 5
group wall union low up
group flow union hydrogen oxygen nanoparticle
pair_style lj/charmm/coul/long 2.5 3.2
bond_style harmonic
angle_style charmm
bond_coeff 1 529.581 1.00
angle_coeff 1 300.0 107.0 50.0 3.0
pair_coeff * * 1.0 1.0
pair_modify mix arithmetic
kspace_style pppm 1e-4
initial velocities
compute mamad flow temp/profile 1.1 0 0 y 20
velocity all create 0.0 12345678 dist uniform
fix 1 all nve
#fix 2 all shake 1e-6 500 0 m 1.0 a 1
fix pressup up aveforce 0.0 0.0 0.0
fix pressdown low aveforce 0.0 0.0 0.0
Couette flow
velocity up set 3.0 0.0 0.0 units box
velocity low set 0.0 0.0 0.0 units box
fix 3 wall setforce 0.0 0.0 0.0
fix 4 flow addforce 0.0 0.0 0.0
fix 5 wall spring/self 57.1
Run
timestep 0.0001
thermo 5000
thermo_style multi
thermo_modify temp mamad
minimize 1.0e-6 1.0e-6 1000 10000
dump 2 all xyz 1 dump.nanofsmooth.XYZ
run 10000
excuse me every one for Previous Incomplete email…