Dear all,
I am trying to stabilize a cross nanorod system of anatase. However during equilibration atoms are getting off the nanorods (screenshots attached for reference). It would be of great help if somebody can explain the reason behind it.
Following is the input script:
#--------------initialization
dimension 3
units metal
atom_style full
newton on
boundary p s p
#-----------system
region simbox block -300 300 -300 300 -100 300
create_box 2 simbox
read_data nr1.xyz add append
read_data nr2.xyz add append
set type 1 charge 2.196
set type 2 charge -1.098
#------------interatomic potential
pair_style buck/coul/cut 8.0
pair_coeff 1 1 31393.4364 0.154 5.2929
pair_coeff 1 2 17106.4193 0.194 12.7031
pair_coeff 2 2 11886.2131 0.234 30.4873
neighbor 2.0 bin
neigh_modify every 1 delay 0 check yes
timestep 0.001
thermo 100
thermo_style custom step cpu temp ke pe etotal pyy lx ly lz
dump config all custom 200 anatase.config.* id type x y z vx vy vz
dump_modify config format line “%8d %5d %25.18g %25.18g %25.18g %20.13g %20.13g %20.13g”
velocity all create 300 12345 mom yes rot no
#------------NVT
fix 2 all nvt temp 300.0 300.0 1.0
run 50000
unfix 2
#----------NPT
fix 1 all npt temp 300.0 300.0 0.1 x 1.0 1.0 1.0 z 1.0 1.0 1.0
run 50000
unfix 1
#------------NVE
fix 3 all nve
run 50000
unfix 3
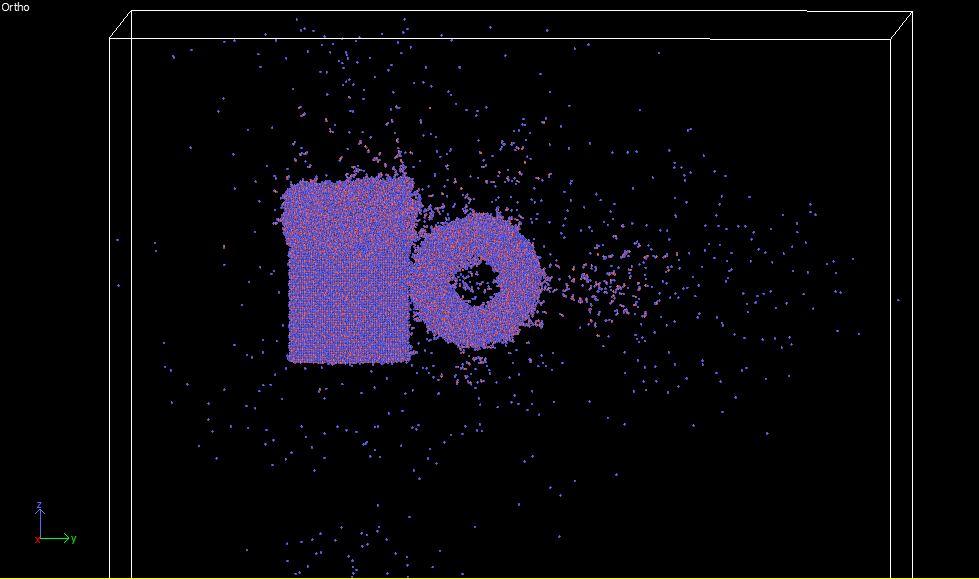
Dear all,
I am trying to stabilize a cross nanorod system of anatase. However during
equilibration atoms are getting off the nanorods (screenshots attached for
reference). It would be of great help if somebody can explain the reason
behind it.
the reason is your input. LAMMPS will read and execute the computations
(e.g. evaluate the forces and update velocities and positions) for the atom
positions and force field parameters you provide it with.
what you see is what happens for the particular settings, geometry and
parameters you input.
so now you have to ask yourself (and run the corresponding tests):
- is the geometry correct? what happens, if you use just one read_data
command instead of two?
- is the force field correct? are the parameters suitable to model a system
like yours (or only bulk crystals where atoms cannot "escape")?
- is your relaxation/equilibration/simulation protocol correct?
it is very unlikely, that somebody here has the time to look into it, and
it *is* part of the job of doing simulations to test, validate and debug
all input and settings. basically, for as long as LAMMPS is doing what it
says in the documentation, it should be doing, then any problem is your
problem. if there is a conflict or contradiction, then the mailing list is
a place to ask people for help.
a couple of obvious things as a starting point to consider:
a) most people do a minimization first; and b) there is no point in using a
barostat on such a system.
axel.
