Dear all,
I am running uniaxial tension deformation. From the dump file, I noticed the order of atoms is rearranged somehow and very different from the original read_data file. Does anyone know why? and also how can I retain the atom order from the read_data file? Thanks a lot. Any suggestion would be appreciated.
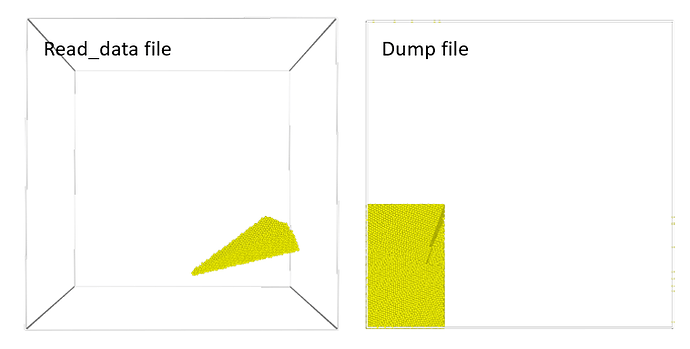
The pictures below show the initial order first 6000 atoms from the read_data file and the order dumped by LAMMPS.
Atoms are distributed across MPI ranks because of the domain decomposition parallelization and then occasionally sorted according to their proximity to each other for efficiency reasons.
When writing dump files, atoms are by default output in the order of the MPI ranks and in the order they are stored in memory for efficiency reasons. That is not a problem in most cases, since the atoms are uniquely identified by their atom ID and thus can be reordered accordingly when reading them.
If you rather waste your valuable simulation time instead and have your atoms resorted every time you output them, you can look into the sort keyword of the dump_modify command — LAMMPS documentation
Hi Alex
Thanks for the response.
I tried using ‘dump_modify sort id’. But for some reason, the order of atoms is still written in the order of the MPI ranks as you mentioned. So I thought maybe the order has already been rewritten when LAMMPS read it, I also tried to modify my read_data to read_data add merge. It still did not retain the original order. Some key information is stored in the original order, so it would be very helpful for me to retain it.
Thanks again for the help. I really appreciated it.
Take care,
Han
Atom IDs have to be unique and if you are using them correctly in your data files and use the dump_modify command correctly, you will get output sorted by the atom ID.
If your atom IDs are not ordered in the way required by your model, that this is a problem of the input your created, not LAMMPS.
Yes, this is exactly what’s confusing me.
I should be able to retain the order by doing so. Please see below the key commands that involve atom id.
Blockquote
region whole block 0 300 0 300 0 300 units box
create_box 3 whole
read_data myStructure.lmp add merge
Blockquote
delete_atoms overlap 2 all all
reset_atom_ids sort yes
Blockquote
dump 1 all custom 10000 dump/tensile*.dump id type x y z
dump_modify 1 sort 1
Just take the “melt” example input from the LAMMPS distribution.
If I run this on 4 CPUs and add these three lines:
dump 1 all custom 10 dump-unsorted.lammpstr id type x y z
dump 2 all custom 10 dump-sorted.lammpstr id type x y z
dump_modify 2 sort 1
I get two different trajectory files where only in the second the lines are sorted by atom ID.
So LAMMPS does what it promises to do. But since I don’t know about your data file and also you are using reset_atom_ids, it is difficult to say where your mistake is. It is not LAMMPS’s fault, though.