I have now tried on two different computers with three different LAMMPS versions, one of them freshly compiled from the develop branch: I get segmentation faults with the inputs attached below and would be thankful for any guidance on what might be the issue (yes, my initial input structure is rather suboptimal, yet I would not expect it to lead to a segmentation fault).
atomistic_pdms.in
# atomistic simulation of PDMS according to https://pubs.acs.org/doi/full/10.1021/ma035760j
# units real
################################ the force-field
# label all types to get a better interface
labelmap atom 1 H 2 C 3 O 4 Si 5 CH3
labelmap bond 1 Si-O 2 Si-CH3
labelmap angle 1 Si-O-Si 2 O-Si-O 3 CH3-Si-CH3 4 O-Si-CH3
labelmap dihedral 1 Si-O-Si-O 2 Si-O-Si-CH3
# Van der Waals & Coulomb interactions / non-bonded
kspace_style pppm 0.00001
# NOTE: the ppm's accuracy and the cut-off for the coul/long are not given in the paper
pair_style hybrid/overlay lj/cut 12.0 lj96/cut 12.0 coul/long 12.0 # angstrom
# the 12-6 potential for interactions involving CH3,
# the 9-6 potential for interactions involving Si & O,
pair_coeff * * lj96/cut 0.0 0.0
pair_coeff * * lj/cut 0.0 0.0
pair_coeff Si Si lj96/cut 0.131 4.29 # ε_αβ [kcal/mol], σ_αβ [Å]
pair_coeff Si O lj96/cut 0.0772 3.94
pair_coeff O O lj96/cut 0.08 3.3
pair_coeff Si CH3 lj/cut 0.1596 3.83
pair_coeff O CH3 lj/cut 0.1247 3.38
pair_coeff CH3 CH3 lj/cut 0.1944 3.73
# Coulomb for all interactions
pair_coeff * * coul/long
# bonded interactions
bond_style harmonic
bond_coeff Si-O 350.12 1.64 # kbond [kcal/(mol Å2)], r0 [Å]
bond_coeff Si-CH3 189.65 1.9
# angles
angle_style harmonic
angle_coeff Si-O-Si 14.14 146.46 # kbend [kcal/(mol rad2)], θ0 [deg]
angle_coeff O-Si-O 94.5 107.82
angle_coeff CH3-Si-CH3 49.97 109.24
angle_coeff O-Si-CH3 49.97 110.69
# dihedrals
dihedral_style harmonic
dihedral_coeff Si-O-Si-O 0.225 1 1 # ktors [kcal/mol], d, n
dihedral_coeff Si-O-Si-CH3 0.01 1 3
################################
# use different time-steps for different forces
run_style respa 3 2 2 bond 1 angle 2 improper 2 dihedral 2 pair 3
timestep 8 # for the outermost respa loop
################################
# thermostat
fix 1 all langevin 400 400 100 5330 # 400 K, gamma = 0.01/fs
minimize_write.in
atom_style hybrid charge molecular
units real
read_data atomistic_melt_pdms_1000_a_20.structure.out
include atomistic_pdms.in
thermo 1
minimize 1.0e-8 1.0e-10 10000 100000
# This is where I get a segmentation fault:
write_data atomistic_melt_pdms_1000_a_20_min.structure.out
thermo 10
run 1000
The atomistic_melt_pdms_1000_a_20.structure.out file can be found here (as it is too large to upload/copy here): https://1drv.ms/u/s!ApWOYAQVCQzhgZeEFrVZl1jhzEdxs7o?e=gbriEM
The output I get
Loop time of 300.051 on 1 procs for 202 steps with 81000 atoms
98.3% CPU use with 1 MPI tasks x 1 OpenMP threads
Minimization stats:
Stopping criterion = quadratic factors are zero
Energy initial, next-to-last, final =
5.90627359709414e+18 1531892637.55781 1537352545.24059
Force two-norm initial, final = 3.5020635e+20 1745034.1
Force max component initial, final = 1.4528833e+20 137280.93
Final line search alpha, max atom move = 6.6254261e-19 9.0954468e-14
Iterations, force evaluations = 202 276
MPI task timing breakdown:
Section | min time | avg time | max time |%varavg| %total
---------------------------------------------------------------
Pair | 256.45 | 256.45 | 256.45 | 0.0 | 85.47
Bond | 6.4108 | 6.4108 | 6.4108 | 0.0 | 2.14
Kspace | 25.818 | 25.818 | 25.818 | 0.0 | 8.60
Neigh | 10.342 | 10.342 | 10.342 | 0.0 | 3.45
Comm | 0.22133 | 0.22133 | 0.22133 | 0.0 | 0.07
Output | 0.052178 | 0.052178 | 0.052178 | 0.0 | 0.02
Modify | 0 | 0 | 0 | 0.0 | 0.00
Other | | 0.7611 | | | 0.25
Nlocal: 81000 ave 81000 max 81000 min
Histogram: 1 0 0 0 0 0 0 0 0 0
Nghost: 56543 ave 56543 max 56543 min
Histogram: 1 0 0 0 0 0 0 0 0 0
Neighs: 1.39802e+07 ave 1.39802e+07 max 1.39802e+07 min
Histogram: 1 0 0 0 0 0 0 0 0 0
Total # of neighbors = 13980151
Ave neighs/atom = 172.59446
Ave special neighs/atom = 8.2222222
Neighbor list builds = 22
Dangerous builds = 0
System init for write_data ...
PPPM initialization ...
[1] 46729 segmentation fault lmp -in
This makes no sense. You are computing distances for all pairs of atoms during all pair styles, even if they do not contribute any forces.
Why not use atom style full?
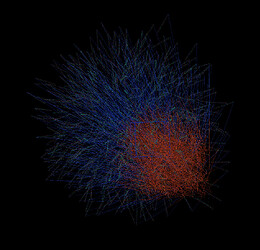
Your initial structure seems bogus. This is what I get when I view it in VMD without and with bonds.
This is confirmed by warnings and extremely higher energies/pressures when running your system:
Step Temp E_pair E_mol TotEng Press
0 0 5.9062736e+18 2.5498022e+09 5.9062736e+18 5.6429238e+17
1 0 9.4368327e+17 2.5498022e+09 9.4368327e+17 8.9695635e+16
2 0 3.1753443e+17 2.5498022e+09 3.1753443e+17 3.0304481e+16
3 0 1.0758831e+17 2.5498022e+09 1.0758831e+17 1.0264289e+16
4 0 3.5728989e+16 2.5498022e+09 3.5728992e+16 3.4098885e+15
5 0 1.3234711e+16 2.5498021e+09 1.3234713e+16 1.2636811e+15
6 0 2.1559453e+15 2.5498018e+09 2.1559479e+15 2.0599899e+14
There are also warnings that should not be there for this kind of system:
WARNING: Inconsistent image flags (src/domain.cpp:815)
WARNING: Bond/angle/dihedral extent > half of periodic box length (src/domain.cpp:936)
I don’t see a segfault, when I run this with the latest develop branch, but there are few more inconsistencies I noticed:
- there is no time integration (I added fix nve)
- you have a mass of 1 assigned to what seems to be a carbon atom
- your r-RESPA sectioning is resulting in too large an inner timestep. with hydrogen atoms, the inner timestep for bonds should be no larger than 0.5fs, better 0.25fs or 0.2fs. but you can do kspace on only every other pair style. Timestep for pair should be no more than 2fs, so timestep would be 4fs. Of course this will only lead to stable time integration after you got rid of the excessive intra-molecular forces.
- in general, if you have a non-equilibrated structure you should start with very conservative settings, use plain verlet time integration and possibly nve/limit to avoid too large displacements and cutoff coulomb to avoid issues with PPPM grids and to speed things up.
Thank you very much for your kind and insightful response. The pair_coeff * * was due to me misunderstanding the pair_style hybrid/overlay, thinking I need coefficients for all combinations. The atom_style hybrid in turn was due to me not having found the atom_style full in the documentation/overlooking it when going over the different atom styles.
I do not actually have any atoms of type one or two, but LAMMPS does not allow a mass of 0 (or a missing mass), that’s why there are the 1s there. That also explains the r-RESPA: without hydrogen, I would expect to be able to use larger time-steps. The paper I cite in the input used: 2 fs time step for the bond length fluctuations, 4 fs time step for the angle, torsion, and improper forces, and an 8 fs time step for the van der Waals forces; maybe I misunderstood the documentation of r-RESPA as well, but I think that’s what I have, no?
I have updated the structure with one that is much better, still a bit bogus (since it starts with the chemically equilibrium bond-lengths, rather than those given by the force-field), but does not lead to any WARNINGs anymore.
I have tried to debug the segfault with lldb and noticed, that it seems to come from some fftw internals. I will try to change my fftw version – as you say that you don’t see it, may I ask what fftw3 version you have installed, @akohlmey?
If you don’t have any atoms of those types, why reserve those types in the first place?
You can confirm the r-RESPA level timestep times from the output.
This is a minor difference and usually does not matter much. The original data seemed to have completely misplaced bonds. The decisive property is the amount of potential energy in the bonded interactions. With just slightly different bond distances, it will decrease quickly during minimization and then stay small as the non-bonded geometry optimization takes place.
I seriously doubt that changing the FFTW version will make a difference. FFTW is one of the most used libraries in scientific computing, it is very mature software, and it has seen only very minor changes for decades and mostly in peripheral features like the MPI wrappers. Besides, LAMMPS only uses FFT libraries at the very simplest level (1d forward and backward FFTs) and does all the 3d FFT related communication and data management with its own wrapper. That itself is one very mature part of LAMMPS and thus also not likely to have issues (in fact, it exists as a standalone package and other projects are using it). If there is any compilation related issue, it is much more likely due to the compiler/OS used than due to the FFT library. If you suspect that there is something wrong, then I would rather suggest to a) try to compile LAMMPS with the built in KISS FFT library (FFT library performance is rarely significant unless you are looking for good strong parallel scaling) and b) try to lower compiler optimization flags.
Please also see my recommendation to start equilibration with cutoff coulomb (and thus bypass FFT issues completely and save time) and plain verlet time integration. Using long-range Coulomb and r-RESPA are only relevant and useful if your system is close to equilibrium and you need accurate forces and want to push for maximum (parallel?) performance.
Thank you again for your comprehensive answers. I have now used the KISS FFT library, with which I did not get anymore segfaults, I can now run my simulations without problems.