Hello Everyone,
Firstly, I use the latest stable version of LAMMPS (02-Aug-23) on Ubuntu 22.04. I conduct a Kremer-Grest simulation of a polymer melt under shear flow and run on the following issue:
At some random timesteps, Rg jumps from an expected value of ~5 to a value of ~1e+3 sigma. Then it goes back to the expected value.
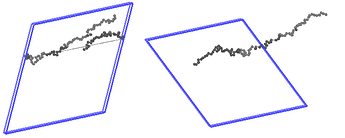
Upon visualization with wrapped coordinates, I find that at those timesteps the chain “escapes” the simulation box. As shown in the attached picture, the chains can appear in both “broken” and “unbroken” states. The former results in the unreasonable Rg values (when I visualize the unwrapped coordinates, I see again the broken chains).
Here is another example where I used as initial snapshot the LAMMPS benchmark (bench/data.chain). Lx should be much bigger, but I am showing this only for visualization purposes. Initially, the chain is wrapped, and then at the next timestep it is made “unbroken” and escapes the box. Once again, at that timestep, Rg diverges.
Further, this problem occurs when I apply higher shear rates. At low shear rates, close to the Newtonian plateau, no such issue is observed. The higher the shear rate, the problem happens at earlier times. The longer the Lx, the issue arises at later times (for the same shear rate).
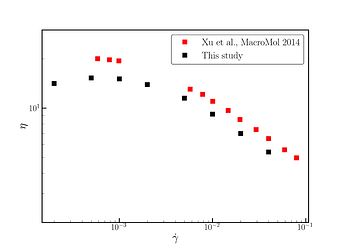
All, the simulations run until complete, and I find that the steady-state viscosity and normal stresses are close to what is available in the literature. There is qualitative, and I guess semiquantititave agreement. However, I use Noser-Hoover thermostat while in the paper (Shear thinning behavior of linear polymer melts under shear flow via nonequilibrium molecular dynamics | The Journal of Chemical Physics | AIP Publishing) dpd thermostat is used, which might explain the differences.
Note that in the last 3 shear rates, the chains have become “broken”.
Some more info:
-
Increased skin distance and communication cutoff (some of these are unnecessary; have tried simpler versions, same problem)
neighbor 1.0 bin
neigh_modify every 1 delay 0 check yes
comm_modify cutoff 3.0 vel yes -
Made sure the system was properly equilibrated and Lx is very large. System dimensions are (8L, L, L) with L = 24.5sigma, density 0.85.
-
To further verify that the initial snapshot is not the cause of the issue, I also used the configuration from LAMMPS benchamarks (bench/in.chain). Same issues were reproduced.
-
Tried Nose-Hoover with 1 and 3 chains.
-
Tried SLLOD and p-SLLOD.
Did not expect anything from the last two steps to happen; were more like desperate attempts to solve the issue. Maybe there is something obvious that I am missing in my simulation. I attach the simulation script, where I have isolated the main commands.
Lastly, what might be the issue is that the box size in the x direction is not long enough, given that the chain expands in the x direction and the pbc creates this weird issue. I can increase the size in the x direction even more, but Lx is already ~ 50Rg(eq.). Any other suggestions? Would the thermostat choice (Nose-Hoover) generate this issue, and dpd would not? I do not see why…
Thank you!
in.lammps (2.0 KB)