Hello everyone,
I am a pharmacist, studying my Master’s Degree in Medical Biotechnology. I am focused on bioinformatics.
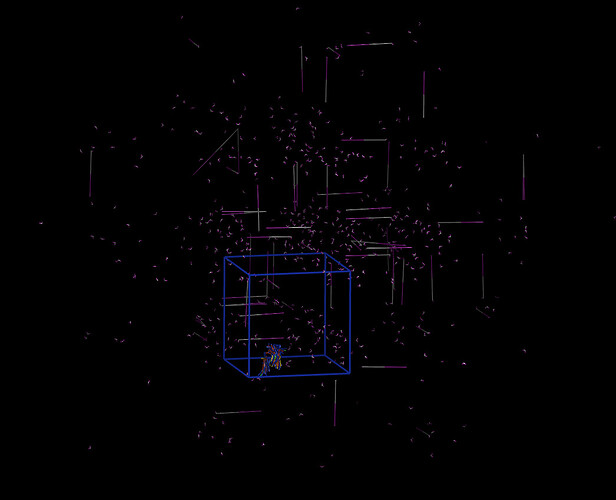
Currently i am working on a project regarding the peptides used in cosmetic preparations and their properties. For this example, i was studying a peptide “Matrixyl” solution and it’s breaking point in thermal stress, and for preparing the simulation i have used the peptide example.
I am sharing with you my data.matrixyl (because of the length kindly access to from my git repo) and the in.peptide file*. I know they are a mass, i just want it to be compiled and get some results (couple of frames for a gif would be perfect).
Also simulation stuck like this:
LAMMPS (3 Mar 2020)
Reading data file ...
orthogonal box = (16.8402 21.0137 9.7681) to (64.2116 68.3851 57.1395)
1 by 1 by 1 MPI processor grid
reading atoms ...
2004 atoms
reading velocities ...
2004 velocities
scanning bonds ...
4 = max bonds/atom
scanning angles ...
6 = max angles/atom
scanning dihedrals ...
15 = max dihedrals/atom
reading bonds ...
1365 bonds
reading angles ...
786 angles
reading dihedrals ...
207 dihedrals
Finding 1-2 1-3 1-4 neighbors ...
special bond factors lj: 0 0 0
special bond factors coul: 0 0 0
4 = max # of 1-2 neighbors
7 = max # of 1-3 neighbors
15 = max # of 1-4 neighbors
19 = max # of special neighbors
special bonds CPU = 0.000424346 secs
read_data CPU = 0.00683676 secs
System init for delete_atoms ...
PPPM initialization ...
WARNING: System is not charge neutral, net charge = 22.967 (src/kspace.cpp:313)
using 12-bit tables for long-range coulomb (src/kspace.cpp:332)
G vector (1/distance) = 0.246118
grid = 20 20 20
stencil order = 5
estimated absolute RMS force accuracy = 0.0319305
estimated relative force accuracy = 9.61578e-05
using double precision FFTW3
3d grid and FFT values/proc = 15625 8000
Neighbor list info ...
update every 1 steps, delay 10 steps, check yes
max neighbors/atom: 2000, page size: 100000
master list distance cutoff = 12
ghost atom cutoff = 12
binsize = 6, bins = 8 8 8
2 neighbor lists, perpetual/occasional/extra = 1 1 0
(1) command delete_atoms, occasional
attributes: full, newton on
pair build: full/bin
stencil: full/bin/3d
bin: standard
(2) pair lj/charmm/coul/long, perpetual
attributes: half, newton on
pair build: half/bin/newton
stencil: half/bin/3d/newton
bin: standard
WARNING: Ignoring 'compress yes' for molecular system (src/delete_atoms.cpp:125)
Deleted 0 atoms, new total = 2004
Finding SHAKE clusters ...
10 = # of size 2 clusters
74 = # of size 3 clusters
0 = # of size 4 clusters
545 = # of frozen angles
find clusters CPU = 0.000286066 secs
131 atoms in group peptide
PPPM initialization ...
using 12-bit tables for long-range coulomb (src/kspace.cpp:332)
G vector (1/distance) = 0.246118
grid = 20 20 20
stencil order = 5
estimated absolute RMS force accuracy = 0.0319305
estimated relative force accuracy = 9.61578e-05
using double precision FFTW3
3d grid and FFT values/proc = 15625 8000
Neighbor list info ...
update every 1 steps, delay 5 steps, check yes
max neighbors/atom: 2000, page size: 100000
master list distance cutoff = 12
ghost atom cutoff = 12
binsize = 6, bins = 8 8 8
1 neighbor lists, perpetual/occasional/extra = 1 0 0
(1) pair lj/charmm/coul/long, perpetual
attributes: half, newton on
pair build: half/bin/newton
stencil: half/bin/3d/newton
bin: standard
Setting up Verlet run ...
Unit style : real
Current step : 0
Time step : 0.5
WARNING: Bond/angle/dihedral extent > half of periodic box length (src/domain.cpp:906)
SHAKE stats (type/ave/delta) on step 0
4 7.37924 4.91615 22
6 7.4382 3.25066 8
18 1.94236 28.2466 3704
31 104.015 158.971
WARNING: Shake determinant < 0.0 (src/RIGID/fix_shake.cpp:1742)
WARNING: Shake determinant < 0.0 (src/RIGID/fix_shake.cpp:1742)
WARNING: Shake determinant < 0.0 (src/RIGID/fix_shake.cpp:1742)
WARNING: Shake determinant < 0.0 (src/RIGID/fix_shake.cpp:1742)
Thank you.