Hi Professor Axel,
I am trying to include the vibrational effect on a binary system A-B for which I have done the cluster expansion. As given in the tutorial, I am using the following command to generate perturbations:
fitsvsl -ms=0.03 -ns=3 -er=5.8
After running the static first-principles calculations, I am fitting using the following command:
fitsvsl -f
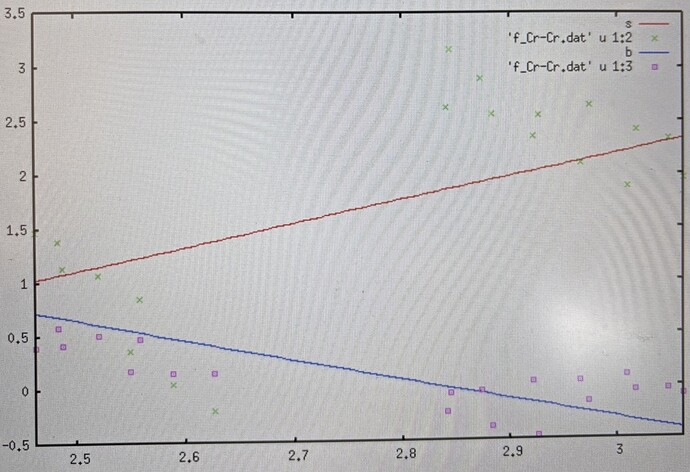
Here something weird is happening in my stiffness vs bond length curve. Whatever value I specify for -ns argument, the fitsvsl.gnu shows 3 chunks of -ns number of data points. Also the bond length varies from 2.5 to 3.0 A. This is similar to fig.2 in CALPHAD 33 (2009) 266-278 (bond length vary over 2.4 to 3A). However there are other references in which bond length varies over a small distance, Fig.1 in Solid State Sciences 41 (2015) 19 - 24 (2.465 to 2.490A), and Figs.3 and 4 in PHYSICAL REVIEW B 85 , 054202 (2012) (2.590 to 2.615A)
I don’t know which is correct. I have checked the frequencies and they are positive. Is there any possible plotting error?
Also it throws error: Unstable modes found. Aborting! for all the structures which are not specified in the strname.in file, when I run the following command:
foreachfile str_relax.out pwd ; svsl -d
I have tried with different combinations of -ns and -er options but I get the same problem.
I don’t understand where I am wrong. Please help.